In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin
- PMID: 34168168
- PMCID: PMC8225877
- DOI: 10.1038/s41598-021-92388-5
In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin
Erratum in
-
Author Correction: In silico comparison of SARS-CoV-2 spike protein-ACE2 binding affinities across species and implications for virus origin.Sci Rep. 2021 Sep 14;11(1):18610. doi: 10.1038/s41598-021-98366-1. Sci Rep. 2021. PMID: 34521997 Free PMC article. No abstract available.
Abstract
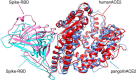
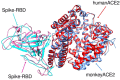
The devastating impact of the COVID-19 pandemic caused by SARS-coronavirus 2 (SARS-CoV-2) has raised important questions about its origins and the mechanism of its transfer to humans. A further question was whether companion or commercial animals could act as SARS-CoV-2 vectors, with early data suggesting susceptibility is species specific. To better understand SARS-CoV-2 species susceptibility, we undertook an in silico structural homology modelling, protein-protein docking, and molecular dynamics simulation study of SARS-CoV-2 spike protein's ability to bind angiotensin converting enzyme 2 (ACE2) from relevant species. Spike protein exhibited the highest binding to human (h)ACE2 of all the species tested, forming the highest number of hydrogen bonds with hACE2. Interestingly, pangolin ACE2 showed the next highest binding affinity despite having a relatively low sequence homology, whereas the affinity of monkey ACE2 was much lower despite its high sequence similarity to hACE2. These differences highlight the power of a structural versus a sequence-based approach to cross-species analyses. ACE2 species in the upper half of the predicted affinity range (monkey, hamster, dog, ferret, cat) have been shown to be permissive to SARS-CoV-2 infection, supporting a correlation between binding affinity and infection susceptibility. These findings show that the earliest known SARS-CoV-2 isolates were surprisingly well adapted to bind strongly to human ACE2, helping explain its efficient human to human respiratory transmission. This study highlights how in silico structural modelling methods can be used to rapidly generate information on novel viruses to help predict their behaviour and aid in countermeasure development.
Conflict of interest statement
The authors declare no competing interests.
Figures

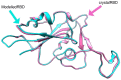
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

