Intratumoral follicular regulatory T cells curtail anti-PD-1 treatment efficacy
- PMID: 34168370
- PMCID: PMC8434898
- DOI: 10.1038/s41590-021-00958-6
Intratumoral follicular regulatory T cells curtail anti-PD-1 treatment efficacy
Abstract
Immune-checkpoint blockade (ICB) has shown remarkable clinical success in boosting antitumor immunity. However, the breadth of its cellular targets and specific mode of action remain elusive. We find that tumor-infiltrating follicular regulatory T (TFR) cells are prevalent in tumor tissues of several cancer types. They are primarily located within tertiary lymphoid structures and exhibit superior suppressive capacity and in vivo persistence as compared with regulatory T cells, with which they share a clonal and developmental relationship. In syngeneic tumor models, anti-PD-1 treatment increases the number of tumor-infiltrating TFR cells. Both TFR cell deficiency and the depletion of TFR cells with anti-CTLA-4 before anti-PD-1 treatment improve tumor control in mice. Notably, in a cohort of 271 patients with melanoma, treatment with anti-CTLA-4 followed by anti-PD-1 at progression was associated with better a survival outcome than monotherapy with anti-PD-1 or anti-CTLA-4, anti-PD-1 followed by anti-CTLA-4 at progression or concomitant combination therapy.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures











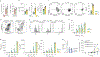

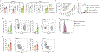
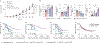
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

