The cellular architecture of the antimicrobial response network in human leprosy granulomas
- PMID: 34168371
- PMCID: PMC8579511
- DOI: 10.1038/s41590-021-00956-8
The cellular architecture of the antimicrobial response network in human leprosy granulomas
Abstract
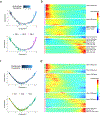
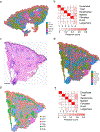
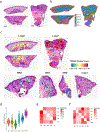
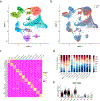
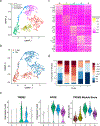
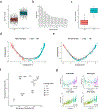
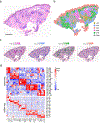
Granulomas are complex cellular structures composed predominantly of macrophages and lymphocytes that function to contain and kill invading pathogens. Here, we investigated the single-cell phenotypes associated with antimicrobial responses in human leprosy granulomas by applying single-cell and spatial sequencing to leprosy biopsy specimens. We focused on reversal reactions (RRs), a dynamic process whereby some patients with disseminated lepromatous leprosy (L-lep) transition toward self-limiting tuberculoid leprosy (T-lep), mounting effective antimicrobial responses. We identified a set of genes encoding proteins involved in antimicrobial responses that are differentially expressed in RR versus L-lep lesions and regulated by interferon-γ and interleukin-1β. By integrating the spatial coordinates of the key cell types and antimicrobial gene expression in RR and T-lep lesions, we constructed a map revealing the organized architecture of granulomas depicting compositional and functional layers by which macrophages, T cells, keratinocytes and fibroblasts can each contribute to the antimicrobial response.
Conflict of interest statement
Declaration of Interests
A.K.S. reports compensation for consulting and/or SAB membership from Merck, Honeycomb Biotechnologies, Cellarity, Cogen Therapeutics, Orche Bio, and Dahlia Biosciences. The remaining authors declare no competing interests.
Figures








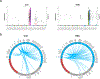

Comment in
-
Spatiotemporal mapping of the leprosy granuloma landscape.Cell Mol Immunol. 2022 May;19(5):558-560. doi: 10.1038/s41423-021-00826-1. Epub 2022 Jan 7. Cell Mol Immunol. 2022. PMID: 34992283 Free PMC article. No abstract available.
References
-
- Gordon S Alternative activation of macrophages. Nat. Rev. Immunol 3, 23–35 (2003). - PubMed
-
- Ridley DS & Jopling WH Classification of leprosy according to immunity. A five-group system. Int J Lepr 34, 255–273 (1966). - PubMed
-
- Modlin RL, Hofman FM, Taylor CR & Rea TH T lymphocyte subsets in the skin lesions of patients with leprosy. J. Am. Acad. Dermatol 8, 182–189 (1983). - PubMed
-
- Stenger S et al. Differential effects of cytolytic T cell subsets on intracellular infection. Science 276, 1684–1687 (1997). - PubMed
-
- Stenger S et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science 282, 121–125 (1998). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

