Biomolecular solid-state NMR spectroscopy at 1200 MHz: the gain in resolution
- PMID: 34170475
- PMCID: PMC8275511
- DOI: 10.1007/s10858-021-00373-x
Biomolecular solid-state NMR spectroscopy at 1200 MHz: the gain in resolution
Abstract
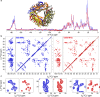
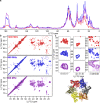
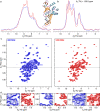
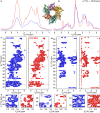
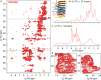
Progress in NMR in general and in biomolecular applications in particular is driven by increasing magnetic-field strengths leading to improved resolution and sensitivity of the NMR spectra. Recently, persistent superconducting magnets at a magnetic field strength (magnetic induction) of 28.2 T corresponding to 1200 MHz proton resonance frequency became commercially available. We present here a collection of high-field NMR spectra of a variety of proteins, including molecular machines, membrane proteins, viral capsids, fibrils and large molecular assemblies. We show this large panel in order to provide an overview over a range of representative systems under study, rather than a single best performing model system. We discuss both carbon-13 and proton-detected experiments, and show that in 13C spectra substantially higher numbers of peaks can be resolved compared to 850 MHz while for 1H spectra the most impressive increase in resolution is observed for aliphatic side-chain resonances.
Keywords: Biomolecular NMR; Helicases; High field; Magic-angle spinning; Solid-state NMR; Viruses.
© 2021. The Author(s).
Figures







References
-
- Abragam A. The principles of nuclear magnetism. Oxford: Oxford Science Publications; 1961.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

