Strain diversity of plant-associated Lactiplantibacillus plantarum
- PMID: 34171185
- PMCID: PMC8449665
- DOI: 10.1111/1751-7915.13871
Strain diversity of plant-associated Lactiplantibacillus plantarum
Abstract
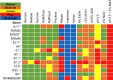
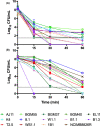
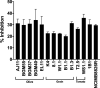
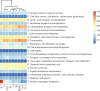
Lactiplantibacillus plantarum (formerly Lactobacillus plantarum) is a lactic acid bacteria species found on plants that is essential for many plant food fermentations. In this study, we investigated the intraspecific phenotypic and genetic diversity of 13 L. plantarum strains isolated from different plant foods, including fermented olives and tomatoes, cactus fruit, teff injera, wheat boza and wheat sourdough starter. We found that strains from the same or similar plant food types frequently exhibited similar carbohydrate metabolism and stress tolerance responses. The isolates from acidic, brine-containing ferments (olives and tomatoes) were more resistant to MRS adjusted to pH 3.5 or containing 4% w/v NaCl, than those recovered from grain fermentations. Strains from fermented olives grew robustly on raffinose as the sole carbon source and were better able to grow in the presence of ethanol (8% v/v or sequential exposure of 8% (v/v) and then 12% (v/v) ethanol) than most isolates from other plant types and the reference strain NCIMB8826R. Cell free culture supernatants from the olive-associated strains were also more effective at inhibiting growth of an olive spoilage strain of Saccharomyces cerevisiae. Multi-locus sequence typing and comparative genomics indicated that isolates from the same source tended to be genetically related. However, despite these similarities, other traits were highly variable between strains from the same plant source, including the capacity for biofilm formation and survival at pH 2 or 50°C. Genomic comparisons were unable to resolve strain differences, with the exception of the most phenotypically impaired and robust isolates, highlighting the importance of utilizing phenotypic studies to investigate differences between strains of L. plantarum. The findings show that L. plantarum is adapted for growth on specific plants or plant food types, but that intraspecific variation may be important for ecological fitness and strain coexistence within individual habitats.
© 2021 The Authors. Microbial Biotechnology published by Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Aquilanti, L., Santarelli, S., Silvestri, G., Osimani, A., Petruzzelli, A., and Clementi, F. (2007) The microbial ecology of a typical Italian salami during its natural fermentation. Int J Food Microbiol 120: 136–145. - PubMed
-
- Barache, N., Ladjouzi, R., Belguesmia, Y., Bendali, F., and Drider, D. (2020) Abundance of Lactobacillus plantarum strains with beneficial attributes in blackberries (Rubus sp.), fresh figs (Ficus carica), and prickly pears (Opuntia ficus‐indica) grown and harvested in Algeria. Probiot Antimicro Protein 12: 1514–1523. - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions

