Directed evolution of potent neutralizing nanobodies against SARS-CoV-2 using CDR-swapping mutagenesis
- PMID: 34171229
- PMCID: PMC8223476
- DOI: 10.1016/j.chembiol.2021.05.019
Directed evolution of potent neutralizing nanobodies against SARS-CoV-2 using CDR-swapping mutagenesis
Abstract
There is widespread interest in facile methods for generating potent neutralizing antibodies, nanobodies, and other affinity proteins against SARS-CoV-2 and related viruses to address current and future pandemics. While isolating antibodies from animals and humans are proven approaches, these methods are limited to the affinities, specificities, and functional activities of antibodies generated by the immune system. Here we report a surprisingly simple directed evolution method for generating nanobodies with high affinities and neutralization activities against SARS-CoV-2. We demonstrate that complementarity-determining region swapping between low-affinity lead nanobodies, which we discovered unintentionally but find is simple to implement systematically, results in matured nanobodies with unusually large increases in affinity. Importantly, the matured nanobodies potently neutralize both SARS-CoV-2 pseudovirus and live virus, and possess drug-like biophysical properties. We expect that our methods will improve in vitro nanobody discovery and accelerate the generation of potent neutralizing nanobodies against diverse coronaviruses.
Keywords: ACE2; COVID-19; RBD; affinity; maturation; nanobody; neutralization; receptor-binding domain; spike; yeast.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




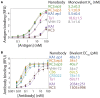


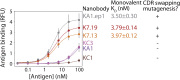
Comment in
-
Facile isolation of high-affinity nanobodies from synthetic libraries using CDR-swapping mutagenesis.STAR Protoc. 2022 Jan 20;3(1):101101. doi: 10.1016/j.xpro.2021.101101. eCollection 2022 Mar 18. STAR Protoc. 2022. PMID: 35098159 Free PMC article.
References
-
- Benatuil L., Perez J.M., Belk J., Hsieh C.-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010;23:155–159. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

