Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: an observational cohort study
- PMID: 34171232
- PMCID: PMC8219489
- DOI: 10.1016/S1473-3099(21)00318-2
Mortality and critical care unit admission associated with the SARS-CoV-2 lineage B.1.1.7 in England: an observational cohort study
Abstract
Background: A more transmissible variant of SARS-CoV-2, the variant of concern 202012/01 or lineage B.1.1.7, has emerged in the UK. We aimed to estimate the risk of critical care admission, mortality in patients who are critically ill, and overall mortality associated with lineage B.1.1.7 compared with non-B.1.1.7. We also compared clinical outcomes between these two groups.
Methods: For this observational cohort study, we linked large primary care (QResearch), national critical care (Intensive Care National Audit & Research Centre Case Mix Programme), and national COVID-19 testing (Public Health England) databases. We used SARS-CoV-2 positive samples with S-gene molecular diagnostic assay failure (SGTF) as a proxy for the presence of lineage B.1.1.7. We extracted two cohorts from the data: the primary care cohort, comprising patients in primary care with a positive community COVID-19 test reported between Nov 1, 2020, and Jan 26, 2021, and known SGTF status; and the critical care cohort, comprising patients admitted for critical care with a positive community COVID-19 test reported between Nov 1, 2020, and Jan 27, 2021, and known SGTF status. We explored the associations between SARS-CoV-2 infection with and without lineage B.1.1.7 and admission to a critical care unit (CCU), 28-day mortality, and 28-day mortality following CCU admission. We used Royston-Parmar models adjusted for age, sex, geographical region, other sociodemographic factors (deprivation index, ethnicity, household housing category, and smoking status for the primary care cohort; and ethnicity, body-mass index, deprivation index, and dependency before admission to acute hospital for the CCU cohort), and comorbidities (asthma, chronic obstructive pulmonary disease, type 1 and 2 diabetes, and hypertension for the primary care cohort; and cardiovascular disease, respiratory disease, metastatic disease, and immunocompromised conditions for the CCU cohort). We reported information on types and duration of organ support for the B.1.1.7 and non-B.1.1.7 groups.
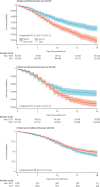
Findings: The primary care cohort included 198 420 patients with SARS-CoV-2 infection. Of these, 117 926 (59·4%) had lineage B.1.1.7, 836 (0·4%) were admitted to CCU, and 899 (0·4%) died within 28 days. The critical care cohort included 4272 patients admitted to CCU. Of these, 2685 (62·8%) had lineage B.1.1.7 and 662 (15·5%) died at the end of critical care. In the primary care cohort, we estimated adjusted hazard ratios (HRs) of 2·15 (95% CI 1·75-2·65) for CCU admission and 1·65 (1·36-2·01) for 28-day mortality for patients with lineage B.1.1.7 compared with the non-B.1.1.7 group. The adjusted HR for mortality in critical care, estimated with the critical care cohort, was 0·91 (0·76-1·09) for patients with lineage B.1.1.7 compared with those with non-B.1.1.7 infection.
Interpretation: Patients with lineage B.1.1.7 were at increased risk of CCU admission and 28-day mortality compared with patients with non-B.1.1.7 SARS-CoV-2. For patients receiving critical care, mortality appeared to be independent of virus strain. Our findings emphasise the importance of measures to control exposure to and infection with COVID-19.
Funding: Wellcome Trust, National Institute for Health Research Oxford Biomedical Research Centre, and the Medical Sciences Division of the University of Oxford.
Copyright © 2021 The Author(s). Published by Elsevier Ltd. This is an Open Access article under the CC BY 4.0 license. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Competing interests JH-C reports receiving grants from the Wellcome trust, Health Data Research-UK, National Institute for Health Research (NIHR) Biomedical Research Centre (Oxford), John Fell Oxford University Press Research Fund, Cancer Research UK, and Oxford Wellcome Institutional Strategic Support Fund; being a member of SAGE subgroups on ethnicity and chair of the NERVTAG risk stratification subgroup; being an unpaid director of QResearch; and being a founder and former director of ClinRisk, outside the submitted work. PST reports previous consultations with AstraZeneca and Duke-National University of Singapore, outside the submitted work. PW was Chief Medical Officer for Sensyne Health, his department received research funding from Sensyne Health, and he holds shares in the company; and he received grant funding from the National Institute for Health Research and Wellcome Trust.
Figures


References
-
- Rambaut A, Loman N, Pybus O. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2021. https://virological.org/t/preliminary-genomic-characterisation-of-an-eme...
-
- Wise J. COVID-19: New coronavirus variant is identified in UK. BMJ. 2020;371 - PubMed
-
- Davies N, Abbott S, Barnard RC. Estimated transmissibility and severity of novel SARS-CoV-2 Variant of Concern 202012/01 in England. 2021. https://cmmid.github.io/topics/covid19/uk-novel-variant.html
-
- Public Health England Investigation of a novel SAR-COVD-2 variant; variant of concern. 2020. https://assets.publishing.service.gov.uk/government/uploads/system/uploa...
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

