Co-occurring gain-of-function mutations in HER2 and HER3 modulate HER2/HER3 activation, oncogenesis, and HER2 inhibitor sensitivity
- PMID: 34171264
- PMCID: PMC8355076
- DOI: 10.1016/j.ccell.2021.06.001
Co-occurring gain-of-function mutations in HER2 and HER3 modulate HER2/HER3 activation, oncogenesis, and HER2 inhibitor sensitivity
Abstract
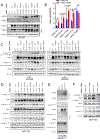
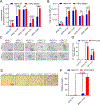
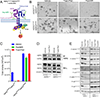
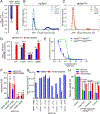
Activating mutations in HER2 (ERBB2) drive the growth of a subset of breast and other cancers and tend to co-occur with HER3 (ERBB3) missense mutations. The HER2 tyrosine kinase inhibitor neratinib has shown clinical activity against HER2-mutant tumors. To characterize the role of HER3 mutations in HER2-mutant tumors, we integrate computational structural modeling with biochemical and cell biological analyses. Computational modeling predicts that the frequent HER3E928G kinase domain mutation enhances the affinity of HER2/HER3 and reduces binding of HER2 to its inhibitor neratinib. Co-expression of mutant HER2/HER3 enhances HER2/HER3 co-immunoprecipitation and ligand-independent activation of HER2/HER3 and PI3K/AKT, resulting in enhanced growth, invasiveness, and resistance to HER2-targeted therapies, which can be reversed by combined treatment with PI3Kα inhibitors. Our results provide a mechanistic rationale for the evolutionary selection of co-occurring HER2/HER3 mutations and the recent clinical observations that HER3 mutations are associated with a poor response to neratinib in HER2-mutant cancers.
Keywords: HER2; HER3; PI3K; Rosetta; breast cancer; molecular dynamics; neratinib; personalized structural biology; precision oncology.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests A.B.H. receives or has received research grant support from Takeda and Lilly and travel support from Puma Biotechnology. J.H. is an employee of Foundation Medicine. A.S.L. is an employee of and holds ownership interest (including patents) in Puma Biotechnology, Inc. C.L.A. receives or has received research grant support from Pfizer, Lilly, Radius, Bayer, and Takeda, holds stock options in Provista, and serves or has served in a scientific advisory role to Puma Biotechnology, Novartis, Lilly, TAIHO Oncology, Daiichi Sankyo, Merck, AstraZeneca, OrigiMed, Immunomedics, Athenex, Arvinas, and the Susan G. Komen Foundation. All other authors declare no competing interests.
Figures



Comment in
-
Mutant HER2 needs mutant HER3 to be an effective oncogene.Cell Rep Med. 2021 Aug 6;2(8):100361. doi: 10.1016/j.xcrm.2021.100361. eCollection 2021 Aug 17. Cell Rep Med. 2021. PMID: 34467245 Free PMC article.
References
-
- Agus DB, Akita RW, Fox WD, Lewis GD, Higgins B, Pisacane PI, Lofgren JA, Tindell C, Evans DP, Maiese K, et al. (2002). Targeting ligand-activated ErbB2 signaling inhibits breast and prostate tumor growth. Cancer Cell 2, 127–137. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

