Inter and intraspecies comparison of the level of selected bacterial phyla in in cattle and sheep based on feces
- PMID: 34172061
- PMCID: PMC8235250
- DOI: 10.1186/s12917-021-02922-w
Inter and intraspecies comparison of the level of selected bacterial phyla in in cattle and sheep based on feces
Abstract
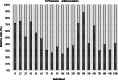
Background: The microbiome of the digestive tract of ruminants contains microbial ecosystem that is affected by both environmental and genetic factors. The subject of this study concerns the influence of selected genetic factors, such as species of animals and "host" individual differences on the digestive tract microbiome composition. The results show the core microbiological composition (Firmicutes and Bacteroidetes) of ruminants digestive tract (based on feces) depending on breed and "host". The Bacteroidetes and Firmicutes phyla are the most abundant in ruminants digestive tract. The aim of the study was to determine the differences prevalence level of Bacteroidetes and Firmicutes phyla in feces of Charolaise cattle and Polish Olkuska Sheep with respect to intra- and inter-species variability.
Results: The research group in the experiment consisted of animals at the age of 3 months kept in the same environmental conditions - rams of Polish Olkuska Sheep (n = 10) and Charolaise bulls (n = 10). Feces were collected individually from each animal (animals without disease symptoms were selected), living on the same environmental conditions. The analysis of the results in terms of species showed differences in the Firmicutes phylum level and Lactobacillaceae family between rams and bulls. Subsequently, the analysis performed for the "host effect" showed differentiation in the levels of the Bacteroidetes and Firmicutes phyla between individuals in a group and also between the groups.
Conclusion: The obtained results suggest that, apart from the diet and the environment, the species and the individual host are equally important factors influencing the microbiological composition of the digestive system of ruminants.
Keywords: Cattle; Digestive tract; Genetic factors; Microbiome; Ruminants; Sheep.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
A preliminary study on farmed and free-ranging mouflons core microbiome.Sci Rep. 2025 Jun 4;15(1):19555. doi: 10.1038/s41598-025-05443-w. Sci Rep. 2025. PMID: 40467971 Free PMC article.
-
Influence of selected factors on the Firmicutes, Bacteroidetes phyla and the Lactobacillaceae family in the digestive tract of sheep.Sci Rep. 2021 Dec 10;11(1):23801. doi: 10.1038/s41598-021-03207-w. Sci Rep. 2021. PMID: 34893656 Free PMC article.
-
Evaluation of Changes in the Levels of Firmicutes and Bacteroidetes Phyla of Sheep Feces Depending on the Breed.Animals (Basel). 2020 Oct 16;10(10):1901. doi: 10.3390/ani10101901. Animals (Basel). 2020. PMID: 33081312 Free PMC article.
-
Microbiome and metabolic disease: revisiting the bacterial phylum Bacteroidetes.J Mol Med (Berl). 2017 Jan;95(1):1-8. doi: 10.1007/s00109-016-1492-2. Epub 2016 Nov 29. J Mol Med (Berl). 2017. PMID: 27900395 Free PMC article. Review.
-
Impact of selected environmental factors on microbiome of the digestive tract of ruminants.BMC Vet Res. 2021 Jan 9;17(1):25. doi: 10.1186/s12917-021-02742-y. BMC Vet Res. 2021. PMID: 33419429 Free PMC article. Review.
Cited by
-
Microbial-Metabolomic Exploration of Tea Polyphenols in the Regulation of Serum Indicators, Liver Metabolism, Rumen Microorganisms, and Metabolism in Hu Sheep.Animals (Basel). 2024 Sep 12;14(18):2661. doi: 10.3390/ani14182661. Animals (Basel). 2024. PMID: 39335251 Free PMC article.
-
Relationship between rumen microbial differences and traits among Hu sheep, Tan sheep, and Dorper sheep.J Anim Sci. 2022 Sep 1;100(9):skac261. doi: 10.1093/jas/skac261. J Anim Sci. 2022. PMID: 35953151 Free PMC article.
-
Metagenomics-Metabolomics Exploration of Three-Way-Crossbreeding Effects on Rumen to Provide Basis for Crossbreeding Improvement of Sheep Microbiome and Metabolome of Sheep.Animals (Basel). 2024 Aug 3;14(15):2256. doi: 10.3390/ani14152256. Animals (Basel). 2024. PMID: 39123781 Free PMC article.
-
Rumen Development of Tianhua Mutton Sheep Was Better than That of Gansu Alpine Fine Wool Sheep under Grazing Conditions.Animals (Basel). 2024 Apr 23;14(9):1259. doi: 10.3390/ani14091259. Animals (Basel). 2024. PMID: 38731263 Free PMC article.
-
Levels of Main Bacterial Phyla in the Gastrointestinal Tract of Sheep Depending on Parity and Age.Animals (Basel). 2021 Jul 25;11(8):2203. doi: 10.3390/ani11082203. Animals (Basel). 2021. PMID: 34438660 Free PMC article.
References
-
- Khafipour E, Li S, Tun H, Derakhshani H, Moossavi S, Plaizier J. Effects of grain feeding on microbiota in the digestive tract of cattle. Anim Front. 2016;6:13–19. doi: 10.2527/af.2016-0018. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources