Dynamics of Verticillium dahliae race 1 population under managed agricultural ecosystems
- PMID: 34172070
- PMCID: PMC8235872
- DOI: 10.1186/s12915-021-01061-w
Dynamics of Verticillium dahliae race 1 population under managed agricultural ecosystems
Abstract
Background: Plant pathogens and their hosts undergo adaptive changes in managed agricultural ecosystems, by overcoming host resistance, but the underlying genetic adaptations are difficult to determine in natural settings. Verticillium dahliae is a fungal pathogen that causes Verticillium wilt on many economically important crops including lettuce. We assessed the dynamics of changes in the V. dahliae genome under selection in a long-term field experiment.
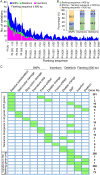
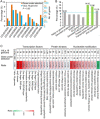
Results: In this study, a field was fumigated before the Verticillium dahliae race 1 strain (VdLs.16) was introduced. A derivative 145-strain population was collected over a 6-year period from this field in which a seggregating population of lettuce derived from Vr1/vr1 parents were evaluated. We de novo sequenced the parental genome of VdLs.16 strain and resequenced the derivative strains to analyze the genetic variations that accumulate over time in the field cropped with lettuce. Population genomics analyses identified 2769 single-nucleotide polymorphisms (SNPs) and 750 insertion/deletions (In-Dels) in the 145 isolates compared with the parental genome. Sequence divergence was identified in the coding sequence regions of 378 genes and in the putative promoter regions of 604 genes. Five-hundred and nine SNPs/In-Dels were identified as fixed. The SNPs and In-Dels were significantly enriched in the transposon-rich, gene-sparse regions, and in those genes with functional roles in signaling and transcriptional regulation.
Conclusions: Under the managed ecosystem continuously cropped to lettuce, the local adaptation of V. dahliae evolves at a whole genome scale to accumulate SNPs/In-Dels nonrandomly in hypervariable regions that encode components of signal transduction and transcriptional regulation.
Keywords: Genetic selection; Local adaptation; Managed agricultural ecosystems; Signal transduction; Transcriptional regulation; Transposon enrichment; Verticillium dahliae.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Genome Sequence of Verticillium dahliae Race 1 Isolate VdLs.16 From Lettuce.Mol Plant Microbe Interact. 2020 Nov;33(11):1265-1269. doi: 10.1094/MPMI-04-20-0103-A. Epub 2020 Sep 24. Mol Plant Microbe Interact. 2020. PMID: 32967552
-
The LsVe1L allele provides a molecular marker for resistance to Verticillium dahliae race 1 in lettuce.BMC Plant Biol. 2019 Jul 10;19(1):305. doi: 10.1186/s12870-019-1905-9. BMC Plant Biol. 2019. PMID: 31291883 Free PMC article.
-
Functional Genomics and Comparative Lineage-Specific Region Analyses Reveal Novel Insights into Race Divergence in Verticillium dahliae.Microbiol Spectr. 2021 Dec 22;9(3):e0111821. doi: 10.1128/Spectrum.01118-21. Epub 2021 Dec 22. Microbiol Spectr. 2021. PMID: 34937170 Free PMC article.
-
An Overview of the Molecular Genetics of Plant Resistance to the Verticillium Wilt Pathogen Verticillium dahliae.Int J Mol Sci. 2020 Feb 7;21(3):1120. doi: 10.3390/ijms21031120. Int J Mol Sci. 2020. PMID: 32046212 Free PMC article. Review.
-
The secretome of Verticillium dahliae in collusion with plant defence responses modulates Verticillium wilt symptoms.Biol Rev Camb Philos Soc. 2022 Oct;97(5):1810-1822. doi: 10.1111/brv.12863. Epub 2022 Apr 27. Biol Rev Camb Philos Soc. 2022. PMID: 35478378 Free PMC article. Review.
Cited by
-
An Efficient Homologous Recombination-Based In Situ Protein-Labeling Method in Verticillium dahliae.Biology (Basel). 2024 Jan 28;13(2):81. doi: 10.3390/biology13020081. Biology (Basel). 2024. PMID: 38392300 Free PMC article.
References
-
- Kaltz O, Shykoff JA. Local adaptation in host–parasite systems. Heredity. 1998;81(4):361–370. doi: 10.1046/j.1365-2540.1998.00435.x. - DOI

