Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis
- PMID: 34172072
- PMCID: PMC8229759
- DOI: 10.1186/s12943-021-01372-0
Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis
Abstract
Background: Colorectal cancer (CRC) is one of the most common malignant tumours. The recurrence and metastasis of CRC seriously affect the survival rate of patients. Angiogenesis is an extremely important cause of tumour growth and metastasis. Circular RNAs (circRNAs) have been emerged as vital regulators for tumour progression. However, the regulatory role, clinical significance and underlying mechanisms still remain largely unknown.
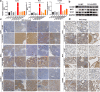
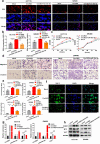
Methods: High-throughput sequencing was used to analyse differential circRNAs expression in tumour and non-tumour tissues of CRC. In situ hybridization (ISH) and qRT-PCR were used to determine the level of circ3823 in CRC tissues and serum samples. Then, functional experiments in vitro and in vivo were performed to investigate the effects of circ3823 on tumour growth, metastasis and angiogenesis in CRC. Sanger sequencing, RNase R and Actinomycin D assay were used to verify the ring structure of circ3823. Mechanistically, dual luciferase reporter assay, fluorescent in situ hybridization (FISH), RNA immunoprecipitation (RIP) and RNA pull-down experiments were performed to confirm the underlying mechanisms of circ3823.
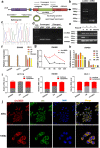
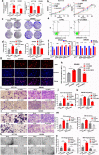
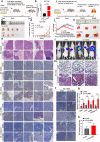
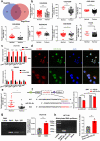
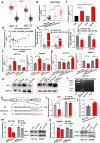
Results: Circ3823 was evidently highly expressed in CRC and high circ3823 expression predicted a worse prognosis of CRC patients. Receiver operating characteristic curves (ROCs) indicated that the expression of circ3823 in serum showed high sensitivity and specificity for detecting CRC which means circ3823 have the potential to be used as diagnostic biomarkers. Functional experiments in vitro and in vivo indicated that circ3823 promote CRC cell proliferation, metastasis and angiogenesis. Mechanism analysis showed that circ3823 act as a competing endogenous RNA of miR-30c-5p to relieve the repressive effect of miR-30c-5p on its target TCF7 which upregulates MYC and CCND1, and finally facilitates CRC progression. In addition, we found that N6-methyladenosine (m6A) modification exists on circ3823. And the m6A modification is involved in regulating the degradation of circ3823.
Conclusions: Our findings suggest that circ3823 promotes CRC growth, metastasis and angiogenesis through circ3823/miR-30c-5p/TCF7 axis and it may serve as a new diagnostic marker or target for treatment of CRC patients. In addition, m6A modification is involved in regulating the degradation of circ3823.
Keywords: Angiogenesis; Colorectal cancer (CRC); N6-methyladenosine (m6A); Tumour progression; circ3823.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

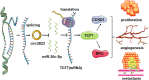
References
-
- Collaborators GBDCC. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet Gastroenterol Hepatol. 2019;4(12):913–933. doi: 10.1016/S2468-1253(19)30345-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

