SCAPP: an algorithm for improved plasmid assembly in metagenomes
- PMID: 34172093
- PMCID: PMC8228940
- DOI: 10.1186/s40168-021-01068-z
SCAPP: an algorithm for improved plasmid assembly in metagenomes
Abstract
Background: Metagenomic sequencing has led to the identification and assembly of many new bacterial genome sequences. These bacteria often contain plasmids: usually small, circular double-stranded DNA molecules that may transfer across bacterial species and confer antibiotic resistance. These plasmids are generally less studied and understood than their bacterial hosts. Part of the reason for this is insufficient computational tools enabling the analysis of plasmids in metagenomic samples.
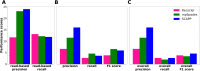
Results: We developed SCAPP (Sequence Contents-Aware Plasmid Peeler)-an algorithm and tool to assemble plasmid sequences from metagenomic sequencing. SCAPP builds on some key ideas from the Recycler algorithm while improving plasmid assemblies by integrating biological knowledge about plasmids. We compared the performance of SCAPP to Recycler and metaplasmidSPAdes on simulated metagenomes, real human gut microbiome samples, and a human gut plasmidome dataset that we generated. We also created plasmidome and metagenome data from the same cow rumen sample and used the parallel sequencing data to create a novel assessment procedure. Overall, SCAPP outperformed Recycler and metaplasmidSPAdes across this wide range of datasets.
Conclusions: SCAPP is an easy to use Python package that enables the assembly of full plasmid sequences from metagenomic samples. It outperformed existing metagenomic plasmid assemblers in most cases and assembled novel and clinically relevant plasmids in samples we generated such as a human gut plasmidome. SCAPP is open-source software available from: https://github.com/Shamir-Lab/SCAPP . Video abstract.
Keywords: Assembly; Plasmids.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Recycler: an algorithm for detecting plasmids from de novo assembly graphs.Bioinformatics. 2017 Feb 15;33(4):475-482. doi: 10.1093/bioinformatics/btw651. Bioinformatics. 2017. PMID: 28003256 Free PMC article.
-
3CAC: improving the classification of phages and plasmids in metagenomic assemblies using assembly graphs.Bioinformatics. 2022 Sep 16;38(Suppl_2):ii56-ii61. doi: 10.1093/bioinformatics/btac468. Bioinformatics. 2022. PMID: 36124804
-
PlasClass improves plasmid sequence classification.PLoS Comput Biol. 2020 Apr 3;16(4):e1007781. doi: 10.1371/journal.pcbi.1007781. eCollection 2020 Apr. PLoS Comput Biol. 2020. PMID: 32243433 Free PMC article.
-
Assessment of metagenomic assemblers based on hybrid reads of real and simulated metagenomic sequences.Brief Bioinform. 2020 May 21;21(3):777-790. doi: 10.1093/bib/bbz025. Brief Bioinform. 2020. PMID: 30860572 Free PMC article. Review.
-
MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices.Methods. 2016 Jun 1;102:3-11. doi: 10.1016/j.ymeth.2016.02.020. Epub 2016 Mar 21. Methods. 2016. PMID: 27012178 Review.
Cited by
-
Recovering Escherichia coli Plasmids in the Absence of Long-Read Sequencing Data.Microorganisms. 2021 Jul 28;9(8):1613. doi: 10.3390/microorganisms9081613. Microorganisms. 2021. PMID: 34442692 Free PMC article.
-
Plasmids of the urinary microbiota.Access Microbiol. 2022 Nov 30;4(11):acmi000429. doi: 10.1099/acmi.0.000429. eCollection 2022. Access Microbiol. 2022. PMID: 36644432 Free PMC article.
-
PlasCAT: Plasmid Cloud Assembly Tool.Bioinformatics. 2024 May 2;40(5):btae299. doi: 10.1093/bioinformatics/btae299. Bioinformatics. 2024. PMID: 38696761 Free PMC article.
-
Plasmids in the human gut reveal neutral dispersal and recombination that is overpowered by inflammatory diseases.Nat Commun. 2024 Apr 11;15(1):3147. doi: 10.1038/s41467-024-47272-x. Nat Commun. 2024. PMID: 38605009 Free PMC article.
-
Precise genotyping of circular mobile elements from metagenomic data uncovers human-associated plasmids with recent common ancestors.Genome Res. 2022 May;32(5):986-1003. doi: 10.1101/gr.275894.121. Epub 2022 Apr 12. Genome Res. 2022. PMID: 35414589 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

