Prediction and validation of hematopoietic stem and progenitor cell off-target editing in transplanted rhesus macaques
- PMID: 34174439
- PMCID: PMC8753565
- DOI: 10.1016/j.ymthe.2021.06.016
Prediction and validation of hematopoietic stem and progenitor cell off-target editing in transplanted rhesus macaques
Abstract
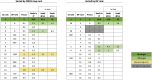
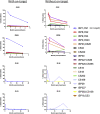
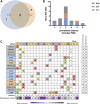
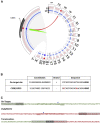
The programmable nuclease technology CRISPR-Cas9 has revolutionized gene editing in the last decade. Due to the risk of off-target editing, accurate and sensitive methods for off-target characterization are crucial prior to applying CRISPR-Cas9 therapeutically. Here, we utilized a rhesus macaque model to compare the predictive values of CIRCLE-seq, an in vitro off-target prediction method, with in silico prediction (ISP) based solely on genomic sequence comparisons. We use AmpliSeq HD error-corrected sequencing to validate off-target sites predicted by CIRCLE-seq and ISP for a CD33 guide RNA (gRNA) with thousands of off-target sites predicted by ISP and CIRCLE-seq. We found poor correlation between the sites predicted by the two methods. When almost 500 sites predicted by each method were analyzed by error-corrected sequencing of hematopoietic cells following transplantation, 19 off-target sites revealed insertion or deletion mutations. Of these sites, 8 were predicted by both methods, 8 by CIRCLE-seq only, and 3 by ISP only. The levels of cells with these off-target edits exhibited no expansion or abnormal behavior in vivo in animals followed for up to 2 years. In addition, we utilized an unbiased method termed CAST-seq to search for translocations between the on-target site and off-target sites present in animals following transplantation, detecting one specific translocation that persisted in blood cells for at least 1 year following transplantation. In conclusion, neither CIRCLE-seq or ISP predicted all sites, and a combination of careful gRNA design, followed by screening for predicted off-target sites in target cells by multiple methods, may be required for optimizing safety of clinical development.
Keywords: CRISPR; Ca9; Macaque; error-corrected sequencing; gene editing; gene therapy; off-target; translocation.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors have no conflicts of interest.
Figures





References
-
- Koo T., Kim J.-S. Therapeutic applications of CRISPR RNA-guided genome editing. Brief. Funct. Genomics. 2017;16:38–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

