Massively parallel sequencing and capillary electrophoresis of a novel panel of falcon STRs: Concordance with minisatellite DNA profiles from historical wildlife crime
- PMID: 34174583
- PMCID: PMC8430417
- DOI: 10.1016/j.fsigen.2021.102550
Massively parallel sequencing and capillary electrophoresis of a novel panel of falcon STRs: Concordance with minisatellite DNA profiles from historical wildlife crime
Abstract
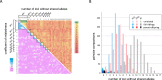
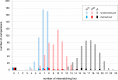
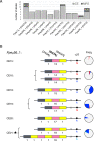
Birds of prey have suffered persecution for centuries through trapping, shooting, poisoning and theft from the wild to meet the demand from egg collectors and falconers; they were also amongst the earliest beneficiaries of DNA testing in wildlife forensics. Here we report the identification and characterisation of 14 novel tetramer, pentamer and hexamer short tandem repeat (STR) markers which can be typed either by capillary electrophoresis or massively parallel sequencing (MPS) and apply them to historical casework samples involving 49 peregrine falcons, 30 of which were claimed to be the captively bred offspring of nine pairs. The birds were initially tested in 1994 with a multilocus DNA fingerprinting probe, a sex test and eight single-locus minisatellite probes (SLPs) demonstrating that 23 birds were unrelated to the claimed parents. The multilocus and SLP approaches were highly discriminating but extremely time consuming and required microgram quantities of high molecular weight DNA and the use of radioisotopes. The STR markers displayed between 2 and 21 alleles per locus (mean = 7.6), lengths between 140 and 360 bp, and heterozygosities from 0.4 to 0.93. They produced wholly concordant conclusions with similar discrimination power but in a fraction of the time using a hundred-fold less DNA and with standard forensic equipment. Furthermore, eleven of these STRs were amplified in a single reaction and typed using MPS on the Illumina MiSeq platform revealing eight additional alleles (three with variant repeat structures and five solely due to flanking SNPs) across four loci. This approach gave a random match probability of < 1E-9, and a parental pair false inclusion probability of < 1E-5, with a further ten-fold reduction in the amount of DNA required (~3 ng) and the potential to analyse mixed samples. These STRs will be of value in monitoring wild populations of these key indicator species as well as for testing captive breeding claims and establishing a database of captive raptors. They have the potential to resolve complex cases involving trace, mixed and degraded samples from raptor persecution casework representing a significant advance over the previously applied methods.
Keywords: DNA forensics; Massively parallel sequencing; Peregrine; Raptor persecution; STR multiplex; Wildlife laundering.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
None.
Figures

Similar articles
-
Mixture deconvolution by massively parallel sequencing of microhaplotypes.Int J Legal Med. 2019 May;133(3):719-729. doi: 10.1007/s00414-019-02010-7. Epub 2019 Feb 13. Int J Legal Med. 2019. PMID: 30758713
-
Developmental validation of the MiSeq FGx Forensic Genomics System for Targeted Next Generation Sequencing in Forensic DNA Casework and Database Laboratories.Forensic Sci Int Genet. 2017 May;28:52-70. doi: 10.1016/j.fsigen.2017.01.011. Epub 2017 Jan 27. Forensic Sci Int Genet. 2017. PMID: 28171784
-
Massively parallel sequencing of forensic STRs and SNPs using the Illumina® ForenSeq™ DNA Signature Prep Kit on the MiSeq FGx™ Forensic Genomics System.Forensic Sci Int Genet. 2017 Nov;31:135-148. doi: 10.1016/j.fsigen.2017.09.003. Epub 2017 Sep 8. Forensic Sci Int Genet. 2017. PMID: 28938154
-
Forensic molecular biomarkers for mixture analysis.Forensic Sci Int Genet. 2019 Jul;41:107-119. doi: 10.1016/j.fsigen.2019.04.003. Epub 2019 Apr 30. Forensic Sci Int Genet. 2019. PMID: 31071519 Review.
-
Massively parallel sequencing techniques for forensics: A review.Electrophoresis. 2018 Nov;39(21):2642-2654. doi: 10.1002/elps.201800082. Epub 2018 Aug 22. Electrophoresis. 2018. PMID: 30101986 Free PMC article. Review.
Cited by
-
Recent advances in forensic biology and forensic DNA typing: INTERPOL review 2019-2022.Forensic Sci Int Synerg. 2022 Dec 27;6:100311. doi: 10.1016/j.fsisyn.2022.100311. eCollection 2023. Forensic Sci Int Synerg. 2022. PMID: 36618991 Free PMC article.
-
An Introductory Overview of Open-Source and Commercial Software Options for the Analysis of Forensic Sequencing Data.Genes (Basel). 2021 Oct 29;12(11):1739. doi: 10.3390/genes12111739. Genes (Basel). 2021. PMID: 34828345 Free PMC article. Review.
-
Sequencing the orthologs of human autosomal forensic short tandem repeats provides individual- and species-level identification in African great apes.BMC Ecol Evol. 2024 Oct 31;24(1):134. doi: 10.1186/s12862-024-02324-0. BMC Ecol Evol. 2024. PMID: 39482599 Free PMC article.
-
Development and validation of a novel 133-plex forensic STR panel (52 STRs and 81 Y-STRs) using single-end 400 bp massive parallel sequencing.Int J Legal Med. 2022 Mar;136(2):447-464. doi: 10.1007/s00414-021-02738-1. Epub 2021 Nov 6. Int J Legal Med. 2022. PMID: 34741666
References
-
- Andreassen R., Schregel J., Kopatz A., Tobiassen C., Knappskog P.M., Hagen S.B., Kleven O., Schneider M., Kojola I., Aspi J., Rykov A. A forensic DNA profiling system for Northern European brown bears (Ursus arctos) Forensic Sci. Int. Genet. 2012;6(6):798–809. - PubMed
-
- Beasley J., Wetton J.H., May C.A. Bird of prey CE and MPS multiplexes: high discrimination for forensic and conservation applications. Forensic Sci. Int. Genet. Suppl. Ser. 2019;7(1):567–568.
-
- Brown J.W., Van Coeverden de Groot P.J., Birt T.P., Seutin G., Boag P.T., Friesen V.L. Appraisal of the consequences of the DDT‐induced bottleneck on the level and geographic distribution of neutral genetic variation in Canadian peregrine falcons, Falco peregrinus. Mol. Ecol. 2007;16(2):327–343. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

