PlasForest: a homology-based random forest classifier for plasmid detection in genomic datasets
- PMID: 34174810
- PMCID: PMC8236179
- DOI: 10.1186/s12859-021-04270-w
PlasForest: a homology-based random forest classifier for plasmid detection in genomic datasets
Abstract
Background: Plasmids are mobile genetic elements that often carry accessory genes, and are vectors for horizontal transfer between bacterial genomes. Plasmid detection in large genomic datasets is crucial to analyze their spread and quantify their role in bacteria adaptation and particularly in antibiotic resistance propagation. Bioinformatics methods have been developed to detect plasmids. However, they suffer from low sensitivity (i.e., most plasmids remain undetected) or low precision (i.e., these methods identify chromosomes as plasmids), and are overall not adapted to identify plasmids in whole genomes that are not fully assembled (contigs and scaffolds).
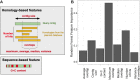
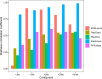
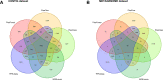
Results: We developed PlasForest, a homology-based random forest classifier identifying bacterial plasmid sequences in partially assembled genomes. Without knowing the taxonomical origin of the samples, PlasForest identifies contigs as plasmids or chromosomes with a F1 score of 0.950. Notably, it can detect 77.4% of plasmid contigs below 1 kb with 2.8% of false positives and 99.9% of plasmid contigs over 50 kb with 2.2% of false positives.
Conclusions: PlasForest outperforms other currently available tools on genomic datasets by being both sensitive and precise. The performance of PlasForest on metagenomic assemblies are currently well below those of other k-mer-based methods, and we discuss how homology-based approaches could improve plasmid detection in such datasets.
Keywords: Genomic datasets; Homology; Plasmid identification; Random forest classifier.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Johnson TJ, Logue CM, Johnson JR, Kuskowski MA, Sherwood JS, Barnes HJ, et al. Associations between multidrug resistance, plasmid content, and virulence potential among extraintestinal pathogenic and commensal Escherichia coli from humans and poultry. Foodborne Pathog Dis. 2012;9:37–46. doi: 10.1089/fpd.2011.0961. - DOI - PMC - PubMed
-
- Heuer H, Binh CTT, Jechalke S, Kopmann C, Zimmerling U, Krögerrecklenfort E, et al. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbiol. 2012 doi: 10.3389/fmicb.2012.00002. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

