Application of machine learning in bacteriophage research
- PMID: 34174831
- PMCID: PMC8235560
- DOI: 10.1186/s12866-021-02256-5
Application of machine learning in bacteriophage research
Abstract
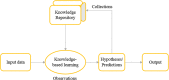
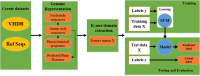
Phages are one of the key components in the structure, dynamics, and interactions of microbial communities in different bins. It has a clear impact on human health and the food industry. Bacteriophage characterization using in vitro approaches are time/cost consuming and laborious tasks. On the other hand, with the advent of new high-throughput sequencing technology, the development of a powerful computational framework to characterize the newly identified bacteriophages is inevitable for future research. Machine learning includes powerful techniques that enable the analysis of complex datasets for knowledge discovery and pattern recognition. In this study, we have conducted a comprehensive review of machine learning methods application using different types of features were applied in various aspects of bacteriophage research including, automated curation, identification, classification, host species recognition, virion protein identification, and life cycle prediction. Moreover, potential limitations and advantages of the developed frameworks were discussed.
Keywords: Bacteriophage; Classification; Host; Life cycle; Machine learning.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Similar articles
-
Global overview and major challenges of host prediction methods for uncultivated phages.Curr Opin Virol. 2021 Aug;49:117-126. doi: 10.1016/j.coviro.2021.05.003. Epub 2021 Jun 12. Curr Opin Virol. 2021. PMID: 34126465 Review.
-
Component Parts of Bacteriophage Virions Accurately Defined by a Machine-Learning Approach Built on Evolutionary Features.mSystems. 2021 Jun 29;6(3):e0024221. doi: 10.1128/mSystems.00242-21. Epub 2021 May 27. mSystems. 2021. PMID: 34042467 Free PMC article.
-
Unveiling the ghost: machine learning's impact on the landscape of virology.J Gen Virol. 2025 Jan;106(1). doi: 10.1099/jgv.0.002067. J Gen Virol. 2025. PMID: 39804261 Review.
-
SourceFinder: a Machine-Learning-Based Tool for Identification of Chromosomal, Plasmid, and Bacteriophage Sequences from Assemblies.Microbiol Spectr. 2022 Dec 21;10(6):e0264122. doi: 10.1128/spectrum.02641-22. Epub 2022 Nov 15. Microbiol Spectr. 2022. PMID: 36377945 Free PMC article.
-
Review and comparative analysis of machine learning-based phage virion protein identification methods.Biochim Biophys Acta Proteins Proteom. 2020 Jun;1868(6):140406. doi: 10.1016/j.bbapap.2020.140406. Epub 2020 Mar 2. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32135196 Review.
Cited by
-
Predictive phage therapy for Escherichia coli urinary tract infections: Cocktail selection for therapy based on machine learning models.Proc Natl Acad Sci U S A. 2024 Mar 19;121(12):e2313574121. doi: 10.1073/pnas.2313574121. Epub 2024 Mar 13. Proc Natl Acad Sci U S A. 2024. PMID: 38478693 Free PMC article.
-
Integrative Systems Biology Analysis Elucidates Mastitis Disease Underlying Functional Modules in Dairy Cattle.Front Genet. 2021 Oct 8;12:712306. doi: 10.3389/fgene.2021.712306. eCollection 2021. Front Genet. 2021. PMID: 34691146 Free PMC article.
-
Genome mining approach reveals the CRISPR-Cas systems features and characteristics in Lactobacillus delbrueckii strains.Heliyon. 2024 Nov 8;10(22):e39920. doi: 10.1016/j.heliyon.2024.e39920. eCollection 2024 Nov 30. Heliyon. 2024. PMID: 39624314 Free PMC article.
-
The CRISPR-Cas system in Lactiplantibacillus plantarum strains: identification and characterization using a genome mining approach.Front Microbiol. 2024 Nov 29;15:1394756. doi: 10.3389/fmicb.2024.1394756. eCollection 2024. Front Microbiol. 2024. PMID: 39678914 Free PMC article.
-
Deep learning-guided structural analysis of a novel bacteriophage KPP105 against multidrug-resistant Klebsiella pneumoniae.Comput Struct Biotechnol J. 2025 May 1;27:1827-1837. doi: 10.1016/j.csbj.2025.04.032. eCollection 2025. Comput Struct Biotechnol J. 2025. PMID: 40470315 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources