Adaptive Resistance Mutations at Suprainhibitory Concentrations Independent of SOS Mutagenesis
- PMID: 34175952
- PMCID: PMC8476149
- DOI: 10.1093/molbev/msab196
Adaptive Resistance Mutations at Suprainhibitory Concentrations Independent of SOS Mutagenesis
Abstract
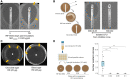
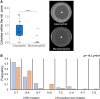
Emergence of resistant bacteria during antimicrobial treatment is one of the most critical and universal health threats. It is known that several stress-induced mutagenesis and heteroresistance mechanisms can enhance microbial adaptation to antibiotics. Here, we demonstrate that the pathogen Bartonella can undergo stress-induced mutagenesis despite the fact it lacks error-prone polymerases, the rpoS gene and functional UV-induced mutagenesis. We demonstrate that Bartonella acquire de novo single mutations during rifampicin exposure at suprainhibitory concentrations at a much higher rate than expected from spontaneous fluctuations. This is while exhibiting a minimal heteroresistance capacity. The emerged resistant mutants acquired a single rpoB mutation, whereas no other mutations were found in their whole genome. Interestingly, the emergence of resistance in Bartonella occurred only during gradual exposure to the antibiotic, indicating that Bartonella sense and react to the changing environment. Using a mathematical model, we demonstrated that, to reproduce the experimental results, mutation rates should be transiently increased over 1,000-folds, and a larger population size or greater heteroresistance capacity is required. RNA expression analysis suggests that the increased mutation rate is due to downregulation of key DNA repair genes (mutS, mutY, and recA), associated with DNA breaks caused by massive prophage inductions. These results provide new evidence of the hazard of antibiotic overuse in medicine and agriculture.
Keywords: Bartonella; antibiotic resistance; evolution; rifampicin; slow-growing bacteria.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures





Similar articles
-
Role of the SOS Response in the Generation of Antibiotic Resistance In Vivo.Antimicrob Agents Chemother. 2021 Jun 17;65(7):e0001321. doi: 10.1128/AAC.00013-21. Epub 2021 Jun 17. Antimicrob Agents Chemother. 2021. PMID: 33875437 Free PMC article.
-
Gamblers: An Antibiotic-Induced Evolvable Cell Subpopulation Differentiated by Reactive-Oxygen-Induced General Stress Response.Mol Cell. 2019 May 16;74(4):785-800.e7. doi: 10.1016/j.molcel.2019.02.037. Epub 2019 Apr 1. Mol Cell. 2019. PMID: 30948267 Free PMC article.
-
RecA and Specialized Error-Prone DNA Polymerases Are Not Required for Mutagenesis and Antibiotic Resistance Induced by Fluoroquinolones in Pseudomonas aeruginosa.Antibiotics (Basel). 2022 Feb 28;11(3):325. doi: 10.3390/antibiotics11030325. Antibiotics (Basel). 2022. PMID: 35326787 Free PMC article.
-
Adaptive mutation and amplification in Escherichia coli: two pathways of genome adaptation under stress.Res Microbiol. 2004 Jun;155(5):352-9. doi: 10.1016/j.resmic.2004.01.020. Res Microbiol. 2004. PMID: 15207867 Review.
-
The central role of the SOS DNA repair system in antibiotics resistance: A new target for a new infectious treatment strategy.Life Sci. 2020 Dec 1;262:118562. doi: 10.1016/j.lfs.2020.118562. Epub 2020 Oct 8. Life Sci. 2020. PMID: 33038378 Review.
Cited by
-
Recent insights into actinobacteria research in antimicrobial resistance: a review.Mol Biol Rep. 2025 Jul 8;52(1):683. doi: 10.1007/s11033-025-10797-5. Mol Biol Rep. 2025. PMID: 40627029 Review.
-
Phenotypic and genotypic characterization of biofilm-producing avian pathogenic Escherichia coli (APEC) isolates from Algerian poultry: associations between antimicrobial resistance and virulence genes.Vet Res Commun. 2025 Jun 21;49(4):232. doi: 10.1007/s11259-025-10801-0. Vet Res Commun. 2025. PMID: 40542899
References
-
- Al Mamun AA, Marians KJ, Humayun MZ.. 2002. DNA polymerase III from Escherichia coli cells expressing mutA mistranslator tRNA is error-prone. J Biol Chem. 277(48):46319–46327. - PubMed
-
- Andrews JM. 2001. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 48(Suppl 1):5–16. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

