Key Considerations for the Development of Safe and Effective SARS-CoV-2 Subunit Vaccine: A Peptide-Based Vaccine Alternative
- PMID: 34176237
- PMCID: PMC8373118
- DOI: 10.1002/advs.202100985
Key Considerations for the Development of Safe and Effective SARS-CoV-2 Subunit Vaccine: A Peptide-Based Vaccine Alternative
Abstract
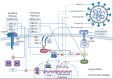
COVID-19 is disastrous to global health and the economy. SARS-CoV-2 infection exhibits similar clinical symptoms and immunopathological sequelae to SARS-CoV infection. Therefore, much of the developmental progress on SARS-CoV vaccines can be utilized for the development of SARS-CoV-2 vaccines. Careful antigen selection during development is always of utmost importance for the production of effective vaccines that do not compromise recipient safety. This holds especially true for SARS-CoV vaccines, as several immunopathological disorders are associated with the activity of structural and nonstructural proteins encoded in the virus's genetic material. Whole viral protein and RNA-encoding full-length proteins contain both protective and "dangerous" sequences, unless pathological fragments are deleted. In light of recent advances, peptide vaccines may present a very safe and effective alternative. Peptide vaccines can avoid immunopathological pro-inflammatory sequences, focus immune responses on neutralizing immunogenic epitopes, avoid off-target antigen loss, combine antigens with different protective roles or mechanisms, even from different viral proteins, and avoid mutant escape by employing highly conserved cryptic epitopes. In this review, an attempt is made to exploit the similarities between SARS-CoV and SARS-CoV-2 in vaccine antigen screening, with particular attention to the pathological and immunogenic properties of SARS proteins.
Keywords: SARS-CoV-2; angiotensin-converting enzyme 2; antibody-dependent enhancement; critical binding residues; neutralizing antibodies; receptor binding domain; spike protein; type-I interferons.
© 2021 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


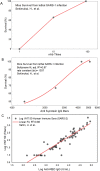
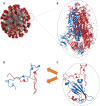
Similar articles
-
Comprehensive characterization of the antibody responses to SARS-CoV-2 Spike protein finds additional vaccine-induced epitopes beyond those for mild infection.Elife. 2022 Jan 24;11:e73490. doi: 10.7554/eLife.73490. Elife. 2022. PMID: 35072628 Free PMC article.
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. Elife. 2020. PMID: 33112236 Free PMC article.
-
Targeting the early life stages of SARS-CoV-2 using a multi-peptide conjugate vaccine.Vaccine. 2025 Apr 30;54:126989. doi: 10.1016/j.vaccine.2025.126989. Epub 2025 Mar 14. Vaccine. 2025. PMID: 40088511
-
Severe acute respiratory syndrome-coronavirus-2 spike (S) protein based vaccine candidates: State of the art and future prospects.Rev Med Virol. 2021 May;31(3):e2183. doi: 10.1002/rmv.2183. Epub 2020 Oct 15. Rev Med Virol. 2021. PMID: 33594794 Free PMC article. Review.
-
Progress and Pitfalls in the Quest for Effective SARS-CoV-2 (COVID-19) Vaccines.Front Immunol. 2020 Oct 2;11:579250. doi: 10.3389/fimmu.2020.579250. eCollection 2020. Front Immunol. 2020. PMID: 33123165 Free PMC article. Review.
Cited by
-
Enhanced Immune Responses by Virus-Mimetic Polymeric Nanostructures Against Infectious Diseases.Front Immunol. 2022 Jan 19;12:804416. doi: 10.3389/fimmu.2021.804416. eCollection 2021. Front Immunol. 2022. PMID: 35126367 Free PMC article. Review.
-
Detection and Quantification of SARS-CoV-2 Receptor Binding Domain Neutralization by a Sensitive Competitive ELISA Assay.Vaccines (Basel). 2021 Dec 16;9(12):1493. doi: 10.3390/vaccines9121493. Vaccines (Basel). 2021. PMID: 34960239 Free PMC article.
-
Kidney Involvement in SARS-CoV-2 Infection: Peritoneal Dialysis as the Preferred Modality.Vaccines (Basel). 2025 Jul 2;13(7):723. doi: 10.3390/vaccines13070723. Vaccines (Basel). 2025. PMID: 40733700 Free PMC article. Review.
-
Non-RBD peptides of SARS-CoV-2 spike protein exhibit immunodominance as they elicit both innate and adaptive immune responses.Heliyon. 2024 Oct 29;10(21):e39941. doi: 10.1016/j.heliyon.2024.e39941. eCollection 2024 Nov 15. Heliyon. 2024. PMID: 39568852 Free PMC article.
-
COVID-19 Variants and Vaccine Development.Viruses. 2024 May 10;16(5):757. doi: 10.3390/v16050757. Viruses. 2024. PMID: 38793638 Free PMC article. Review.
References
-
- Dong E., Du H., Gardner L., Lancet Infect. Dis. 2020, 20, 533. - PubMed
-
- World Health Organization. 2008. Middle East Respiratory Syndrome Corona Virus (MERS‐CoV) Global Summary and Assessment ofRisk, 2018, 8. https://www.who.int/csr/disease/coronavirus_infections/risk‐assessment‐a...
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
