A brief introduction to the analysis of time-series data from biologging studies
- PMID: 34176325
- PMCID: PMC8237163
- DOI: 10.1098/rstb.2020.0227
A brief introduction to the analysis of time-series data from biologging studies
Abstract
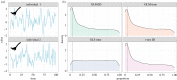
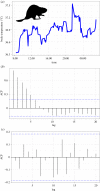
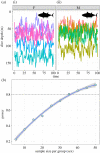
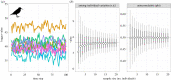
Recent advances in tagging and biologging technology have yielded unprecedented insights into wild animal physiology. However, time-series data from such wild tracking studies present numerous analytical challenges owing to their unique nature, often exhibiting strong autocorrelation within and among samples, low samples sizes and complicated random effect structures. Gleaning robust quantitative estimates from these physiological data, and, therefore, accurate insights into the life histories of the animals they pertain to, requires careful and thoughtful application of existing statistical tools. Using a combination of both simulated and real datasets, I highlight the key pitfalls associated with analysing physiological data from wild monitoring studies, and investigate issues of optimal study design, statistical power, and model precision and accuracy. I also recommend best practice approaches for dealing with their inherent limitations. This work will provide a concise, accessible roadmap for researchers looking to maximize the yield of information from complex and hard-won biologging datasets. This article is part of the theme issue 'Measuring physiology in free-living animals (Part II)'.
Keywords: animal movement; animal physiology; mixed models; temporal autocorrelation; time-series model.
Figures
Similar articles
-
Introduction to the theme issue: Measuring physiology in free-living animals.Philos Trans R Soc Lond B Biol Sci. 2021 Aug 2;376(1830):20200210. doi: 10.1098/rstb.2020.0210. Epub 2021 Jun 14. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34121463 Free PMC article.
-
Animal tag technology keeps coming of age: an engineering perspective.Philos Trans R Soc Lond B Biol Sci. 2021 Aug 16;376(1831):20200229. doi: 10.1098/rstb.2020.0229. Epub 2021 Jun 28. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34176328 Free PMC article.
-
What is physiologging? Introduction to the theme issue, part 2.Philos Trans R Soc Lond B Biol Sci. 2021 Aug 16;376(1831):20210028. doi: 10.1098/rstb.2021.0028. Epub 2021 Jun 28. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34176329 Free PMC article.
-
Biologging and Biotelemetry: Tools for Understanding the Lives and Environments of Marine Animals.Annu Rev Anim Biosci. 2023 Feb 15;11:247-267. doi: 10.1146/annurev-animal-050322-073657. Annu Rev Anim Biosci. 2023. PMID: 36790885 Review.
-
Integrating physiology, behavior, and energetics: Biologging in a free-living arctic hibernator.Comp Biochem Physiol A Mol Integr Physiol. 2016 Dec;202:53-62. doi: 10.1016/j.cbpa.2016.04.020. Epub 2016 Apr 29. Comp Biochem Physiol A Mol Integr Physiol. 2016. PMID: 27139082 Review.
Cited by
-
Introduction to the theme issue: Measuring physiology in free-living animals.Philos Trans R Soc Lond B Biol Sci. 2021 Aug 2;376(1830):20200210. doi: 10.1098/rstb.2020.0210. Epub 2021 Jun 14. Philos Trans R Soc Lond B Biol Sci. 2021. PMID: 34121463 Free PMC article.
-
Temperature and land use change are associated with Rana temporaria reproductive success and phenology.PeerJ. 2024 Aug 30;12:e17901. doi: 10.7717/peerj.17901. eCollection 2024. PeerJ. 2024. PMID: 39224827 Free PMC article.
-
Hemoglobin signal network mapping reveals novel indicators for precision medicine.Sci Rep. 2023 Oct 25;13(1):18257. doi: 10.1038/s41598-023-43694-7. Sci Rep. 2023. PMID: 37880310 Free PMC article.
-
Physiological evidence of stress reduction during a summer Antarctic expedition with a significant influence of previous experience and vigor.Sci Rep. 2024 Feb 17;14(1):3981. doi: 10.1038/s41598-024-54203-9. Sci Rep. 2024. PMID: 38368474 Free PMC article.
-
Time synchronisation for millisecond-precision on bio-loggers.Mov Ecol. 2024 Oct 28;12(1):71. doi: 10.1186/s40462-024-00512-7. Mov Ecol. 2024. PMID: 39468685 Free PMC article.
References
-
- Hawkes LA, Exeter O, Henderson SM, Kerry C, Kukuluya A, Rudd J, Whelan S, Yoder N, Witt MJ. 2020. Autonomous underwater videography and tracking of basking sharks. Anim. Biotelemetry 8, 29. (10.1186/s40317-020-00216-w) - DOI
-
- Smith BJ, Hart KM, Mazzotti FJ, Basille M, Romagosa CM. 2018. Evaluating GPS biologging technology for studying spatial ecology of large constricting snakes. Anim. Biotelemetry 6, 1. (10.1186/s40317-018-0145-3) - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

