Characterizing Network Search Algorithms Developed for Dynamic Causal Modeling
- PMID: 34177506
- PMCID: PMC8222613
- DOI: 10.3389/fninf.2021.656486
Characterizing Network Search Algorithms Developed for Dynamic Causal Modeling
Abstract
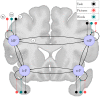
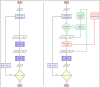
Dynamic causal modeling (DCM) is a widely used tool to estimate the effective connectivity of specified models of a brain network. Finding the model explaining measured data is one of the most important outstanding problems in Bayesian modeling. Using heuristic model search algorithms enables us to find an optimal model without having to define a model set a priori. However, the development of such methods is cumbersome in the case of large model-spaces. We aimed to utilize commonly used graph theoretical search algorithms for DCM to create a framework for characterizing them, and to investigate relevance of such methods for single-subject and group-level studies. Because of the enormous computational demand of DCM calculations, we separated the model estimation procedure from the search algorithm by providing a database containing the parameters of all models in a full model-space. For test data a publicly available fMRI dataset of 60 subjects was used. First, we reimplemented the deterministic bilinear DCM algorithm in the ReDCM R package, increasing computational speed during model estimation. Then, three network search algorithms have been adapted for DCM, and we demonstrated how modifications to these methods, based on DCM posterior parameter estimates, can enhance search performance. Comparison of the results are based on model evidence, structural similarities and the number of model estimations needed during search. An analytical approach using Bayesian model reduction (BMR) for efficient network discovery is already available for DCM. Comparing model search methods we found that topological algorithms often outperform analytical methods for single-subject analysis and achieve similar results for recovering common network properties of the winning model family, or set of models, obtained by multi-subject family-wise analysis. However, network search methods show their limitations in higher level statistical analysis of parametric empirical Bayes. Optimizing such linear modeling schemes the BMR methods are still considered the recommended approach. We envision the freely available database of estimated model-spaces to help further studies of the DCM model-space, and the ReDCM package to be a useful contribution for Bayesian inference within and beyond the field of neuroscience.
Keywords: dynamic causal modeling; fMRI; model-space; network topology; search algorithm.
Copyright © 2021 Aranyi, Nagy, Opposits, Berényi and Emri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
A strategy of model space search for dynamic causal modeling in task fMRI data exploratory analysis.Phys Eng Sci Med. 2022 Sep;45(3):867-882. doi: 10.1007/s13246-022-01156-w. Epub 2022 Jul 18. Phys Eng Sci Med. 2022. PMID: 35849323
-
mpdcm: A toolbox for massively parallel dynamic causal modeling.J Neurosci Methods. 2016 Jan 15;257:7-16. doi: 10.1016/j.jneumeth.2015.09.009. Epub 2015 Sep 16. J Neurosci Methods. 2016. PMID: 26384541
-
A spectral sampling algorithm in dynamic causal modelling for resting-state fMRI.Hum Brain Mapp. 2023 Jun 1;44(8):2981-2992. doi: 10.1002/hbm.26256. Epub 2023 Mar 16. Hum Brain Mapp. 2023. PMID: 36929686 Free PMC article.
-
fMRI in Non-human Primate: A Review on Factors That Can Affect Interpretation and Dynamic Causal Modeling Application.Front Neurosci. 2019 Sep 18;13:973. doi: 10.3389/fnins.2019.00973. eCollection 2019. Front Neurosci. 2019. PMID: 31619951 Free PMC article. Review.
-
An introduction to thermodynamic integration and application to dynamic causal models.Cogn Neurodyn. 2022 Feb;16(1):1-15. doi: 10.1007/s11571-021-09696-9. Epub 2021 Jul 25. Cogn Neurodyn. 2022. PMID: 35116083 Free PMC article. Review.
Cited by
-
A strategy of model space search for dynamic causal modeling in task fMRI data exploratory analysis.Phys Eng Sci Med. 2022 Sep;45(3):867-882. doi: 10.1007/s13246-022-01156-w. Epub 2022 Jul 18. Phys Eng Sci Med. 2022. PMID: 35849323
-
The Constrained Disorder Principle Overcomes the Challenges of Methods for Assessing Uncertainty in Biological Systems.J Pers Med. 2024 Dec 28;15(1):10. doi: 10.3390/jpm15010010. J Pers Med. 2024. PMID: 39852203 Free PMC article. Review.
References
-
- Copeland M., Soh J., Puca A., Manning M., Gollob D. (2015). Microsoft Azure: Planning, Deploying, and Managing Your Data Center in the Cloud, 1st Edn. Berkeley, CA: Apress. 10.1007/978-1-4842-1043-7 - DOI
LinkOut - more resources
Full Text Sources

