Glycyrrhizic Acid for COVID-19: Findings of Targeting Pivotal Inflammatory Pathways Triggered by SARS-CoV-2
- PMID: 34177566
- PMCID: PMC8223069
- DOI: 10.3389/fphar.2021.631206
Glycyrrhizic Acid for COVID-19: Findings of Targeting Pivotal Inflammatory Pathways Triggered by SARS-CoV-2
Abstract
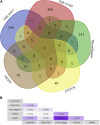
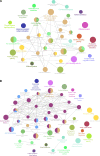
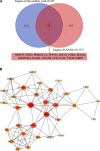
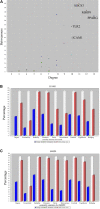
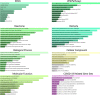
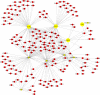
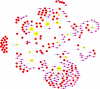
Background: Coronavirus disease 2019 (COVID-19) is now a worldwide public health crisis. The causative pathogen is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Novel therapeutic agents are desperately needed. Because of the frequent mutations in the virus and its ability to cause cytokine storms, targeting the viral proteins has some drawbacks. Targeting cellular factors or pivotal inflammatory pathways triggered by SARS-CoV-2 may produce a broader range of therapies. Glycyrrhizic acid (GA) might be beneficial against SARS-CoV-2 because of its anti-inflammatory and antiviral characteristics and possible ability to regulate crucial host factors. However, the mechanism underlying how GA regulates host factors remains to be determined. Methods: In our report, we conducted a bioinformatics analysis to identify possible GA targets, biological functions, protein-protein interactions, transcription-factor-gene interactions, transcription-factor-miRNA coregulatory networks, and the signaling pathways of GA against COVID-19. Results: Protein-protein interactions and network analysis showed that ICAM1, MMP9, TLR2, and SOCS3 had higher degree values, which may be key targets of GA for COVID-19. GO analysis indicated that the response to reactive oxygen species was significantly enriched. Pathway enrichment analysis showed that the IL-17, IL-6, TNF-α, IFN signals, complement system, and growth factor receptor signaling are the main pathways. The interactions of TF genes and miRNA with common targets and the activity of TFs were also recognized. Conclusions: GA may inhibit COVID-19 through its anti-oxidant, anti-viral, and anti-inflammatory effects, and its ability to activate the immune system, and targeted therapy for those pathways is a predominant strategy to inhibit the cytokine storms triggered by SARS-CoV-2 infection.
Keywords: SARS-CoV-2; coronavirus disease 2019; glycyrrhizic acid; inflammatory pathways; network pharmacology.
Copyright © 2021 Zheng, Huang, Lai, Liu, Jiang and Zhan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19).Gene. 2020 Dec 15;762:145057. doi: 10.1016/j.gene.2020.145057. Epub 2020 Aug 15. Gene. 2020. PMID: 32805314 Free PMC article.
-
Pathogenesis-directed therapy of 2019 novel coronavirus disease.J Med Virol. 2021 Mar;93(3):1320-1342. doi: 10.1002/jmv.26610. Epub 2020 Nov 10. J Med Virol. 2021. PMID: 33073355 Review.
-
Bioinformatics analyses of significant genes, related pathways, and candidate diagnostic biomarkers and molecular targets in SARS-CoV-2/COVID-19.Gene Rep. 2020 Dec;21:100956. doi: 10.1016/j.genrep.2020.100956. Epub 2020 Nov 4. Gene Rep. 2020. PMID: 33553808 Free PMC article.
-
The pharmacological mechanism of Huashi Baidu Formula for the treatment of COVID-19 by combined network pharmacology and molecular docking.Ann Palliat Med. 2021 Apr;10(4):3864-3895. doi: 10.21037/apm-20-1759. Epub 2021 Mar 8. Ann Palliat Med. 2021. PMID: 33691446
-
Disruption of CCR5 signaling to treat COVID-19-associated cytokine storm: Case series of four critically ill patients treated with leronlimab.J Transl Autoimmun. 2021;4:100083. doi: 10.1016/j.jtauto.2021.100083. Epub 2021 Jan 6. J Transl Autoimmun. 2021. PMID: 33521616 Free PMC article. Review.
Cited by
-
Advancing combination treatment with glycyrrhizin and boswellic acids for hospitalized patients with moderate COVID-19 infection: a randomized clinical trial.Inflammopharmacology. 2022 Apr;30(2):477-486. doi: 10.1007/s10787-022-00939-7. Epub 2022 Mar 1. Inflammopharmacology. 2022. PMID: 35233748 Free PMC article. Clinical Trial.
-
Gene network inference from single-cell omics data and domain knowledge for constructing COVID-19-specific ICAM1-associated pathways.Front Genet. 2023 Aug 31;14:1250545. doi: 10.3389/fgene.2023.1250545. eCollection 2023. Front Genet. 2023. PMID: 37719701 Free PMC article.
-
Differential Co-Expression Network Analysis Reveals Key Hub-High Traffic Genes as Potential Therapeutic Targets for COVID-19 Pandemic.Front Immunol. 2021 Dec 15;12:789317. doi: 10.3389/fimmu.2021.789317. eCollection 2021. Front Immunol. 2021. PMID: 34975885 Free PMC article.
-
Glycyrrhizic Acid and Its Derivatives: Promising Candidates for the Management of Type 2 Diabetes Mellitus and Its Complications.Int J Mol Sci. 2022 Sep 20;23(19):10988. doi: 10.3390/ijms231910988. Int J Mol Sci. 2022. PMID: 36232291 Free PMC article. Review.
-
Evaluation of the Antiviral Efficacy of Subcutaneous Nafamostat Formulated with Glycyrrhizic Acid against SARS-CoV-2 in a Murine Model.Int J Mol Sci. 2023 May 31;24(11):9579. doi: 10.3390/ijms24119579. Int J Mol Sci. 2023. PMID: 37298530 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

