Proerythroblast Cells of Diamond-Blackfan Anemia Patients With RPS19 and CECR1 Mutations Have Similar Transcriptomic Signature
- PMID: 34177624
- PMCID: PMC8226250
- DOI: 10.3389/fphys.2021.679919
Proerythroblast Cells of Diamond-Blackfan Anemia Patients With RPS19 and CECR1 Mutations Have Similar Transcriptomic Signature
Abstract
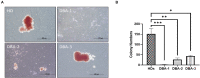
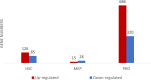
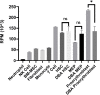
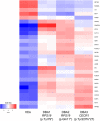
Diamond Blackfan Anemia (DBA) is an inherited bone marrow (BM) failure syndrome, characterized by a paucity of erythroid differentiation. DBA is mainly caused by the mutations in ribosomal protein genes, hence classified as ribosomopathy. However, in approximately 30% of patients, the molecular etiology cannot be discovered. RPS19 germline mutations caused 25% of the cases. On the other hand, CECR1 mutations also cause phenotypes similar to DBA but not being a ribosomopathy. Due to the blockade of erythropoiesis in the BM, we investigated the transcriptomic profile of three different cell types of BM resident cells of DBA patients and compared them with healthy donors. From BM aspirates BM mononuclear cells (MNCs) were isolated and hematopoietic stem cells (HSC) [CD71-CD34+ CD38mo/lo], megakaryocyte-erythroid progenitor cells (MEP) [CD71-CD34+ CD38hi] and Proerythroblasts [CD71+ CD117+ CD38+] were sorted and analyzed with a transcriptomic approach. Among all these cells, proerythroblasts had the most different transcriptomic profile. The genes associated with cellular stress/immune responses were increased and some of the transcription factors that play a role in erythroid differentiation had altered expression in DBA proerythroblasts. We also showed that gene expression levels of ribosomal proteins were decreased in DBA proerythroblasts. In addition to these, colony formation assay (CFU-E) provided functional evidence of the failure of erythroid differentiation in DBA patients. According to our findings that all patients resembling both RPS19 and CECR1 mutations have common transcriptomic signatures, it may be possible that inflammatory BM niche may have a role in DBA pathogenesis.
Keywords: CECR1; Diamond Blackfan Anemia; RPS19; proerythroblast; ribosomopathy; transcriptomics.
Copyright © 2021 Karaosmanoglu, Kursunel, Uckan Cetinkaya, Gumruk, Esendagli, Unal and Taskiran.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Proliferation deficiency of multipotent hematopoietic progenitors in ribosomal protein S19 (RPS19)-deficient diamond-Blackfan anemia improves following RPS19 gene transfer.Mol Ther. 2003 May;7(5 Pt 1):613-22. doi: 10.1016/s1525-0016(03)00091-1. Mol Ther. 2003. PMID: 12718904
-
Gene transfer improves erythroid development in ribosomal protein S19-deficient Diamond-Blackfan anemia.Blood. 2002 Oct 15;100(8):2724-31. doi: 10.1182/blood.V100.8.2724. Blood. 2002. PMID: 12351378
-
An RPS19-edited model for Diamond-Blackfan anemia reveals TP53-dependent impairment of hematopoietic stem cell activity.JCI Insight. 2023 Jan 10;8(1):e161810. doi: 10.1172/jci.insight.161810. JCI Insight. 2023. PMID: 36413407 Free PMC article.
-
Hematopoietic cell transplantation and gene therapy for Diamond-Blackfan anemia: state of the art and science.Front Oncol. 2023 Sep 11;13:1236038. doi: 10.3389/fonc.2023.1236038. eCollection 2023. Front Oncol. 2023. PMID: 37752993 Free PMC article. Review.
-
Recent insights into the pathogenesis of Diamond-Blackfan anaemia.Br J Haematol. 2006 Oct;135(2):149-57. doi: 10.1111/j.1365-2141.2006.06268.x. Epub 2006 Aug 31. Br J Haematol. 2006. PMID: 16942586 Review.
Cited by
-
SATB1 Chromatin Loops Regulate Megakaryocyte/Erythroid Progenitor Expansion by Facilitating HSP70 and GATA1 Induction.Stem Cells. 2023 Jun 15;41(6):560-569. doi: 10.1093/stmcls/sxad025. Stem Cells. 2023. PMID: 36987811 Free PMC article.
-
Downregulation of SATB1 by miRNAs reduces megakaryocyte/erythroid progenitor expansion in preclinical models of Diamond-Blackfan anemia.Exp Hematol. 2022 Jul;111:66-78. doi: 10.1016/j.exphem.2022.04.005. Epub 2022 Apr 20. Exp Hematol. 2022. PMID: 35460833 Free PMC article.
References
-
- Attar A. (2014). Changes in the Cell Surface Markers During Normal Hematopoiesis: A Guide to Cell Isolation. Glob. J. Hematol. Blood Transfus. 1 20–28. 10.15379/2408-9877.2014.01.01.4 - DOI
-
- Avondo F., Roncaglia P., Crescenzio N., Krmac H., Garelli E., Armiraglio M., et al. (2009). Fibroblasts from patients with Diamond-Blackfan anaemia show abnormal expression of genes involved in protein synthesis, amino acid metabolism and cancer. BMC Genomics 10:442. 10.1186/1471-2164-10-442 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

