Comparative Proteomic Analysis of Histoplasma capsulatum Yeast and Mycelium Reveals Differential Metabolic Shifts and Cell Wall Remodeling Processes in the Different Morphotypes
- PMID: 34177824
- PMCID: PMC8226243
- DOI: 10.3389/fmicb.2021.640931
Comparative Proteomic Analysis of Histoplasma capsulatum Yeast and Mycelium Reveals Differential Metabolic Shifts and Cell Wall Remodeling Processes in the Different Morphotypes
Abstract
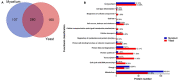
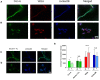
Histoplasma capsulatum is a thermally dimorphic fungus distributed worldwide, but with the highest incidence in the Americas within specific geographic areas, such as the Mississippi River Valley and regions in Latin America. This fungus is the etiologic agent of histoplasmosis, an important life-threatening systemic mycosis. Dimorphism is an important feature for fungal survival in different environments and is related to the virulence of H. capsulatum, and essential to the establishment of infection. Proteomic profiles have made important contributions to the knowledge of metabolism and pathogenicity in several biological models. However, H. capsulatum proteome studies have been underexplored. In the present study, we report the first proteomic comparison between the mycelium and the yeast cells of H. capsulatum. Liquid chromatography coupled to mass spectrometry was used to evaluate the proteomic profile of the two phases of H. capsulatum growth, mycelium, and yeast. In summary, 214 and 225 proteins were only detected/or preferentially abundant in mycelium or yeast cells, respectively. In mycelium, enzymes related to the glycolytic pathway and to the alcoholic fermentation occurred in greater abundance, suggesting a higher use of anaerobic pathways for energy production. In yeast cells, proteins related to the tricarboxylic acid cycle and response to temperature stress were in high abundance. Proteins related to oxidative stress response or involved with cell wall metabolism were identified with differential abundance in both conditions. Proteomic data validation was performed by enzymatic activity determination, Western blot assays, or immunofluorescence microscopy. These experiments corroborated, directly or indirectly, the abundance of isocitrate lyase, 2-methylcitrate synthase, catalase B, and mannosyl-oligosaccharide-1,2-alpha-mannosidase in the mycelium and heat shock protein (HSP) 30, HSP60, glucosamine-fructose-6-phosphate aminotransferase, glucosamine-6-phosphate deaminase, and N-acetylglucosamine-phosphate mutase in yeast cells. The proteomic profile-associated functional classification analyses of proteins provided new and interesting information regarding the differences in metabolism between the two distinct growth forms of H. capsulatum.
Keywords: Histoplasma capsulatum; dimorphism; fungal biology; mycelium; proteomic analysis; yeast.
Copyright © 2021 Almeida, Baeza, Almeida-Paes, Bailão, Borges, Guimarães, Soares and Zancopé-Oliveira.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Almeida M. A., Almeida-Paes R., Guimarães A. J., Valente R. H., Soares C. M. A., Zancopé-Oliveira R. M. (2020). Immunoproteomics reveals pathogen’s antigens involved in Homo sapiens-Histoplasma capsulatum interaction and specific linear B-cell epitopes in histoplasmosis. Front. Cell. Infect. Microbiol. 29:591121. 10.3389/fcimb.2020.591121 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

