A New Ciboria sp. for Soil Mycoremediation and the Bacterial Contribution to the Depletion of Total Petroleum Hydrocarbons
- PMID: 34177829
- PMCID: PMC8221241
- DOI: 10.3389/fmicb.2021.647373
A New Ciboria sp. for Soil Mycoremediation and the Bacterial Contribution to the Depletion of Total Petroleum Hydrocarbons
Abstract
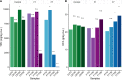
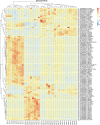
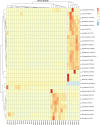
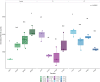
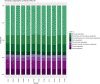
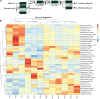
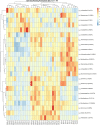
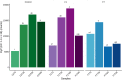
A Ciboria sp. strain (Phylum Ascomycota) was isolated from hydrocarbon-polluted soil of an abandoned oil refinery in Italy. The strain was able to utilize diesel oil as a sole carbon source for growth. Laboratory-scale experiments were designed to evaluate the use of this fungal strain for treatment of the polluted soil. The concentration of total petroleum hydrocarbons (TPH) in the soil was 8,538 mg/kg. Mesocosms containing the contaminated soil were inoculated with the fungal strain at 1 or 7%, on a fresh weight base ratio. After 90 days of incubation, the depletion of TPH contamination was of 78% with the 1% inoculant, and 99% with the 7% inoculant. 16S rDNA and ITS metabarcoding of the bacterial and fungal communities was performed in order to evaluate the potential synergism between fungi and bacteria in the bioremediation process. The functional metagenomic prediction indicated Arthrobacter, Dietzia, Brachybacerium, Brevibacterium, Gordonia, Leucobacter, Lysobacter, and Agrobacterium spp. as generalist saprophytes, essential for the onset of hydrocarbonoclastic specialist bacterial species, identified as Streptomyces, Nocardoides, Pseudonocardia, Solirubrobacter, Parvibaculum, Rhodanobacter, Luteiomonas, Planomicrobium, and Bacillus spp., involved in the TPH depletion. The fungal metabolism accelerated the onset of specialist over generalist bacteria. The capacity of the Ciboria sp. to deplete TPH in the soil in treatment was also ascertained.
Keywords: Ciboria sp.; dye-decolorizing peroxidases DyP; functional metagenomic prediction; generalist bacterial species; mycoremediation; specialist bacterial species; total petroleum hydrocarbons.
Copyright © 2021 Becarelli, Chicca, La China, Siracusa, Bardi, Gullo, Petroni, Levin and Di Gregorio.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Hydrocarbonoclastic Ascomycetes to enhance co-composting of total petroleum hydrocarbon (TPH) contaminated dredged sediments and lignocellulosic matrices.N Biotechnol. 2019 May 25;50:27-36. doi: 10.1016/j.nbt.2019.01.006. Epub 2019 Jan 14. N Biotechnol. 2019. PMID: 30654133
-
Effect of soil organic matter on petroleum hydrocarbon degradation in diesel/fuel oil-contaminated soil.J Biosci Bioeng. 2020 May;129(5):603-612. doi: 10.1016/j.jbiosc.2019.12.001. Epub 2020 Jan 25. J Biosci Bioeng. 2020. PMID: 31992527
-
Bioremediation of diesel and lubricant oil-contaminated soils using enhanced landfarming system.Chemosphere. 2016 Dec;164:558-567. doi: 10.1016/j.chemosphere.2016.08.128. Epub 2016 Sep 11. Chemosphere. 2016. PMID: 27627466
-
The impact of lead co-contamination on ecotoxicity and the bacterial community during the bioremediation of total petroleum hydrocarbon-contaminated soils.Environ Pollut. 2019 Oct;253:939-948. doi: 10.1016/j.envpol.2019.07.107. Epub 2019 Jul 20. Environ Pollut. 2019. PMID: 31351302
-
Fungal bioremediation of soil co-contaminated with petroleum hydrocarbons and toxic metals.Appl Microbiol Biotechnol. 2020 Nov;104(21):8999-9008. doi: 10.1007/s00253-020-10854-y. Epub 2020 Sep 17. Appl Microbiol Biotechnol. 2020. PMID: 32940735 Free PMC article. Review.
Cited by
-
Main Factors Determining the Scale-Up Effectiveness of Mycoremediation for the Decontamination of Aliphatic Hydrocarbons in Soil.J Fungi (Basel). 2023 Dec 16;9(12):1205. doi: 10.3390/jof9121205. J Fungi (Basel). 2023. PMID: 38132804 Free PMC article. Review.
-
Fungal bioproducts for petroleum hydrocarbons and toxic metals remediation: recent advances and emerging technologies.Bioprocess Biosyst Eng. 2023 Mar;46(3):393-428. doi: 10.1007/s00449-022-02763-3. Epub 2022 Aug 9. Bioprocess Biosyst Eng. 2023. PMID: 35943595 Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

