Genomic Characterization of mcr-1.1-Producing Escherichia coli Recovered From Human Infections in São Paulo, Brazil
- PMID: 34177843
- PMCID: PMC8221240
- DOI: 10.3389/fmicb.2021.663414
Genomic Characterization of mcr-1.1-Producing Escherichia coli Recovered From Human Infections in São Paulo, Brazil
Abstract
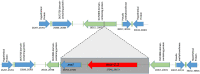
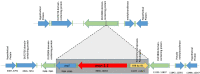
Polymyxins are one of most important antibiotics available for multidrug-resistant Gram-negative infections. Diverse chromosomal resistance mechanisms have been described, but the polymyxin resistance phenotype is not yet completely understood. The objective of this study was to characterize colistin resistant mcr-1-producing strains isolated from human infections over one year in a hospital setting (Hospital das Clínicas, São Paulo, Brazil). We isolated 490 colistin-resistant Gram-negative rods, of which eight were mcr-1.1-positive Escherichia coli, the only species with this result, indicating a low incidence of the mcr-1 production mechanism among colistin-resistant isolates. All mcr-1.1 positive isolates showed similarly low MICs for colistin and were susceptible to most antibiotics tested. The isolates showed diversity of MLST classification. The eight mcr-1.1-positive E. coli genomes were sequenced. In seven of eight isolates the mcr-1.1 gene is located in a contig that is presumed to be a part of an IncX4 plasmid; in one isolate, it is located in a contig that is presumed to be part of an IncHI2A plasmid. Three different genomic contexts for mcr-1.1 were observed, including a genomic cassette mcr-1.1-pap2 disrupting a DUF2806 domain-containing gene in six isolates. In addition, an IS1-family transposase was found inserted next to the mcr-1.1 cassette in one isolate. An mcr-1.1-pap2 genomic cassette not disrupting any gene was identified in another isolate. Our results suggest that plasmid dissemination of hospital-resident strains took place during the study period and highlight the need for continued genomic surveillance.
Keywords: Polymyxin; colistin; hospital dissemination; insertion sequence; plasmid.
Copyright © 2021 Girardello, Piroupo, Martins, Maffucci, Cury, Franco, Malta, Rocha, Pinho, Rossi, Duarte and Setubal.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Agência Nacional de Vigilância Sanitária (2016). Boletim de Segurança do Paciente e Qualidade em Serviços de Saúde N° 14: Avaliação dos Indicadores Nacionais das Infecções Relacionadas à Assistência à Saúde (IRAS) e Resistência Microbiana do Ano de 2015. Brasília: Agência Nacional de Vigilância Sanitária.
-
- Aires C. A. M., da Conceição-Neto O. C., Tavares E., Oliveira T. R., Dias C. F., Montezzi L. F., et al. (2017). Emergence of the plasmid-mediated MCR-1 gene in clinical KPC-2-producing Klebsiella Pneumoniae sequence type 392 in brazil. Antimicrob. Agents Chemother. 61 e317–e317. 10.1128/AAC.00317-17 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

