Comparative Analysis of the IclR-Family of Bacterial Transcription Factors and Their DNA-Binding Motifs: Structure, Positioning, Co-Evolution, Regulon Content
- PMID: 34177859
- PMCID: PMC8222616
- DOI: 10.3389/fmicb.2021.675815
Comparative Analysis of the IclR-Family of Bacterial Transcription Factors and Their DNA-Binding Motifs: Structure, Positioning, Co-Evolution, Regulon Content
Abstract
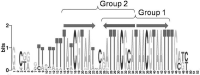
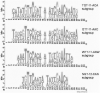
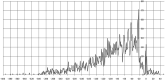
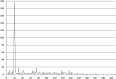
The IclR-family is a large group of transcription factors (TFs) regulating various biological processes in diverse bacteria. Using comparative genomics techniques, we have identified binding motifs of IclR-family TFs, reconstructed regulons and analyzed their content, finding co-occurrences between the regulated COGs (clusters of orthologous genes), useful for future functional characterizations of TFs and their regulated genes. We describe two main types of IclR-family motifs, similar in sequence but different in the arrangement of the half-sites (boxes), with GKTYCRYW3-4RYGRAMC and TGRAACAN1-2TGTTYCA consensuses, and also predict that TFs in 32 orthologous groups have binding sites comprised of three boxes with alternating direction, which implies two possible alternative modes of dimerization of TFs. We identified trends in site positioning relative to the translational gene start, and show that TFs in 94 orthologous groups bind tandem sites with 18-22 nucleotides between their centers. We predict protein-DNA contacts via the correlation analysis of nucleotides in binding sites and amino acids of the DNA-binding domain of TFs, and show that the majority of interacting positions and predicted contacts are similar for both types of motifs and conform well both to available experimental data and to general protein-DNA interaction trends.
Keywords: IclR-family; comparative genomics; protein-DNA contacts; tandem binding sites; transcription factor binding sites (TFBS); transcription regulation.
Copyright © 2021 Suvorova and Gelfand.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
GntR Family of Bacterial Transcription Factors and Their DNA Binding Motifs: Structure, Positioning and Co-Evolution.PLoS One. 2015 Jul 7;10(7):e0132618. doi: 10.1371/journal.pone.0132618. eCollection 2015. PLoS One. 2015. PMID: 26151451 Free PMC article.
-
Comparative genomics of pyridoxal 5'-phosphate-dependent transcription factor regulons in Bacteria.Microb Genom. 2016 Jan 18;2(1):e000047. doi: 10.1099/mgen.0.000047. eCollection 2016 Jan. Microb Genom. 2016. PMID: 28348826 Free PMC article.
-
Comparative genomics and evolution of regulons of the LacI-family transcription factors.Front Microbiol. 2014 Jun 11;5:294. doi: 10.3389/fmicb.2014.00294. eCollection 2014. Front Microbiol. 2014. PMID: 24966856 Free PMC article.
-
Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors.FEMS Microbiol Rev. 2006 Mar;30(2):157-86. doi: 10.1111/j.1574-6976.2005.00008.x. FEMS Microbiol Rev. 2006. PMID: 16472303 Review.
-
The MocR-like transcription factors: pyridoxal 5'-phosphate-dependent regulators of bacterial metabolism.FEBS J. 2018 Nov;285(21):3925-3944. doi: 10.1111/febs.14599. Epub 2018 Jul 5. FEBS J. 2018. PMID: 29974999 Review.
Cited by
-
Molecular mechanism of GylR-mediated regulation of glycerol metabolism in Streptomyces clavuligerus NRRL 3585.Front Microbiol. 2022 Nov 25;13:1078293. doi: 10.3389/fmicb.2022.1078293. eCollection 2022. Front Microbiol. 2022. PMID: 36504789 Free PMC article.
-
Degradation of C19-Steroids and Effect of Androstenedione on Gene Expression in Nocardioides simplex.Curr Microbiol. 2025 Feb 19;82(4):143. doi: 10.1007/s00284-025-04105-4. Curr Microbiol. 2025. PMID: 39969609
-
Tuning the performance of a TphR-based terephthalate biosensor with a design of experiments approach.Metab Eng Commun. 2024 Nov 6;19:e00250. doi: 10.1016/j.mec.2024.e00250. eCollection 2024 Dec. Metab Eng Commun. 2024. PMID: 39618701 Free PMC article.
-
Strategies for Improving Small-Molecule Biosensors in Bacteria.Biosensors (Basel). 2022 Jan 25;12(2):64. doi: 10.3390/bios12020064. Biosensors (Basel). 2022. PMID: 35200325 Free PMC article. Review.
-
Unveiling the regulatory mechanisms of salicylate degradation gene cluster cehGHIR4 in Rhizobium sp. strain X9.Appl Environ Microbiol. 2023 Oct 31;89(10):e0080223. doi: 10.1128/aem.00802-23. Epub 2023 Oct 6. Appl Environ Microbiol. 2023. PMID: 37800922 Free PMC article.
References
-
- Arias-Barrau E., Olivera E. R., Luengo J. M., Fernández C., Galán B., García J. L., et al. (2004). The homogentisate pathway: a central catabolic pathway involved in the degradation of L-phenylalanine, L-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186 5062–5077. 10.1128/jb.186.15.5062-5077.2004 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

