Molecular and Mechanistic Characterization of PddB, the First PLP-Independent 2,4-Diaminobutyric Acid Racemase Discovered in an Actinobacterial D-Amino Acid Homopolymer Biosynthesis
- PMID: 34177872
- PMCID: PMC8225329
- DOI: 10.3389/fmicb.2021.686023
Molecular and Mechanistic Characterization of PddB, the First PLP-Independent 2,4-Diaminobutyric Acid Racemase Discovered in an Actinobacterial D-Amino Acid Homopolymer Biosynthesis
Abstract
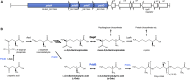
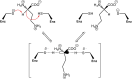
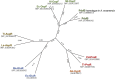
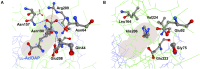
We recently disclosed that the biosynthesis of antiviral γ-poly-D-2,4-diaminobutyric acid (poly-D-Dab) in Streptoalloteichus hindustanus involves an unprecedented cofactor independent stereoinversion of Dab catalyzed by PddB, which shows weak homology to diaminopimelate epimerase (DapF). Enzymological properties and mechanistic details of this enzyme, however, had remained to be elucidated. Here, through a series of biochemical characterizations, structural modeling, and site-directed mutageneses, we fully illustrate the first Dab-specific PLP-independent racemase PddB and further provide an insight into its evolution. The activity of the recombinant PddB was shown to be optimal around pH 8.5, and its other fundamental properties resembled those of typical PLP-independent racemases/epimerases. The enzyme catalyzed Dab specific stereoinversion with a calculated equilibrium constant of nearly unity, demonstrating that the reaction catalyzed by PddB is indeed racemization. Its activity was inhibited upon incubation with sulfhydryl reagents, and the site-directed substitution of two putative catalytic Cys residues led to the abolishment of the activity. These observations provided critical evidence that PddB employs the thiolate-thiol pair to catalyze interconversion of Dab isomers. Despite the low levels of sequence similarity, a phylogenetic analysis of PddB indicated its particular relevance to DapF among PLP-independent racemases/epimerases. Secondary structure prediction and 3D structural modeling of PddB revealed its remarkable conformational analogy to DapF, which in turn allowed us to predict amino acid residues potentially responsible for the discrimination of structural difference between diaminopimelate and its specific substrate, Dab. Further, PddB homologs which seemed to be narrowly distributed only in actinobacterial kingdom were constantly encoded adjacent to the putative poly-D-Dab synthetase gene. These observations strongly suggested that PddB could have evolved from the primary metabolic DapF in order to organize the biosynthesis pathway for the particular secondary metabolite, poly-D-Dab. The present study is on the first molecular characterization of PLP-independent Dab racemase and provides insights that could contribute to further discovery of unprecedented PLP-independent racemases.
Keywords: 2; 4-diaminobutyric acid; D-amino acid; PLP-independent; amino acid racemase; biosynthesis; diaminopimelate epimerase; homo poly-amino acid; secondary metabolite.
Copyright © 2021 Yamanaka, Ozaki, Hamano and Oikawa.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
The Stereocontrolled Biosynthesis of Mirror-Symmetric 2,4-Diaminobutyric Acid Homopolymers Is Critically Governed by Adenylation Activations.ACS Chem Biol. 2020 Jul 17;15(7):1964-1973. doi: 10.1021/acschembio.0c00321. Epub 2020 Jun 16. ACS Chem Biol. 2020. PMID: 32484328
-
Characterization of DcsC, a PLP-independent racemase involved in the biosynthesis of D-cycloserine.Org Biomol Chem. 2012 Mar 21;10(11):2248-54. doi: 10.1039/c2ob06864h. Epub 2012 Feb 6. Org Biomol Chem. 2012. PMID: 22307920
-
Chlamydia trachomatis dapF Encodes a Bifunctional Enzyme Capable of Both d-Glutamate Racemase and Diaminopimelate Epimerase Activities.mBio. 2018 Apr 3;9(2):e00204-18. doi: 10.1128/mBio.00204-18. mBio. 2018. PMID: 29615498 Free PMC article.
-
Catalytic mechanism and properties of pyridoxal 5'-phosphate independent racemases: how enzymes alter mismatched acidity and basicity.Nat Prod Rep. 2019 Dec 11;36(12):1687-1705. doi: 10.1039/c9np00017h. Nat Prod Rep. 2019. PMID: 30994146 Review.
-
Stereospecificity for the hydrogen transfer and molecular evolution of pyridoxal enzymes.Biosci Biotechnol Biochem. 1996 Feb;60(2):181-7. doi: 10.1271/bbb.60.181. Biosci Biotechnol Biochem. 1996. PMID: 9063963 Review.
Cited by
-
Structure and inhibition of diaminopimelic acid epimerase by slow-binding α-methyl amino acids.Protein Sci. 2025 May;34(5):e70139. doi: 10.1002/pro.70139. Protein Sci. 2025. PMID: 40299312 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

