Cross-Tissue Transcriptomic Analysis Leveraging Machine Learning Approaches Identifies New Biomarkers for Rheumatoid Arthritis
- PMID: 34177888
- PMCID: PMC8223752
- DOI: 10.3389/fimmu.2021.638066
Cross-Tissue Transcriptomic Analysis Leveraging Machine Learning Approaches Identifies New Biomarkers for Rheumatoid Arthritis
Abstract
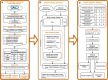
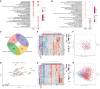
There is an urgent need to identify biomarkers for diagnosis and disease activity monitoring in rheumatoid arthritis (RA). We leveraged publicly available microarray gene expression data in the NCBI GEO database for whole blood (N=1,885) and synovial (N=284) tissues from RA patients and healthy controls. We developed a robust machine learning feature selection pipeline with validation on five independent datasets culminating in 13 genes: TNFAIP6, S100A8, TNFSF10, DRAM1, LY96, QPCT, KYNU, ENTPD1, CLIC1, ATP6V0E1, HSP90AB1, NCL and CIRBP which define the RA score and demonstrate its clinical utility: the score tracks the disease activity DAS28 (p = 7e-9), distinguishes osteoarthritis (OA) from RA (OR 0.57, p = 8e-10) and polyJIA from healthy controls (OR 1.15, p = 2e-4) and monitors treatment effect in RA (p = 2e-4). Finally, the immunoblotting analysis of six proteins on an independent cohort confirmed two proteins, TNFAIP6/TSG6 and HSP90AB1/HSP90.
Keywords: biomarker; blood; gene expression; machine learning; rheumatoid arthritis; synovium.
Copyright © 2021 Rychkov, Neely, Oskotsky, Yu, Perlmutter, Nititham, Carvidi, Krueger, Gross, Criswell, Ashouri and Sirota.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

