Insights Into the Mechanisms Implicated in Pinus pinaster Resistance to Pinewood Nematode
- PMID: 34178007
- PMCID: PMC8222992
- DOI: 10.3389/fpls.2021.690857
Insights Into the Mechanisms Implicated in Pinus pinaster Resistance to Pinewood Nematode
Abstract
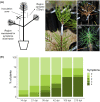
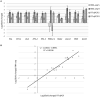
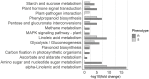
Pine wilt disease (PWD), caused by the plant-parasitic nematode Bursaphelenchus xylophilus, has become a severe environmental problem in the Iberian Peninsula with devastating effects in Pinus pinaster forests. Despite the high levels of this species' susceptibility, previous studies reported heritable resistance in P. pinaster trees. Understanding the basis of this resistance can be of extreme relevance for future programs aiming at reducing the disease impact on P. pinaster forests. In this study, we highlighted the mechanisms possibly involved in P. pinaster resistance to PWD, by comparing the transcriptional changes between resistant and susceptible plants after infection. Our analysis revealed a higher number of differentially expressed genes (DEGs) in resistant plants (1,916) when compared with susceptible plants (1,226). Resistance to PWN is mediated by the induction of the jasmonic acid (JA) defense pathway, secondary metabolism pathways, lignin synthesis, oxidative stress response genes, and resistance genes. Quantification of the acetyl bromide-soluble lignin confirmed a significant increase of cell wall lignification of stem tissues around the inoculation zone in resistant plants. In addition to less lignified cell walls, susceptibility to the pine wood nematode seems associated with the activation of the salicylic acid (SA) defense pathway at 72 hpi, as revealed by the higher SA levels in the tissues of susceptible plants. Cell wall reinforcement and hormone signaling mechanisms seem therefore essential for a resistance response.
Keywords: Bursaphelenchus xylophilus; cell wall lignification; jasmonate; maritime pine; pine wilt disease; resistance genes; secondary metabolism; transcriptome.
Copyright © 2021 Modesto, Sterck, Arbona, Gómez-Cadenas, Carrasquinho, Van de Peer and Miguel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Adie B., Chico J. M., Rubio-Somoza I., Solano R. (2007). Modulation of plant defenses by ethylene. J. Plant Growth Regul. 26, 160–177. 10.1007/s00344-007-0012-6 - DOI
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
LinkOut - more resources
Full Text Sources

