Whole-Exome Sequencing Analysis of Human Semen Quality in Russian Multiethnic Population
- PMID: 34178030
- PMCID: PMC8232892
- DOI: 10.3389/fgene.2021.662846
Whole-Exome Sequencing Analysis of Human Semen Quality in Russian Multiethnic Population
Abstract
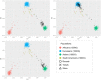
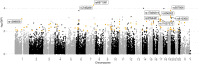
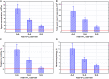
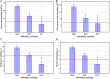
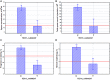
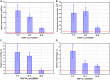
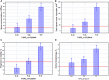
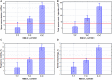
The global trend toward the reduction of human spermatogenic function observed in many countries, including Russia, raised the problem of extensive screening and monitoring of male fertility and elucidation of its genetic and ethnic mechanisms. Recently, whole-exome sequencing (WES) was developed as a powerful tool for genetic analysis of complex traits. We present here the first Russian WES study for identification of new genes associated with semen quality. The experimental 3 × 2 design of the WES study was based on the analysis of 157 samples including three ethnic groups-Slavs (59), Buryats (n = 49), and Yakuts (n = 49), and two different semen quality groups-pathozoospermia (n = 95) and normospermia (n = 62). Additionally, our WES study group was negative for complete AZF microdeletions of the Y-chromosome. The normospermia group included men with normal sperm parameters in accordance with the WHO-recommended reference limit. The pathozoospermia group included men with impaired semen quality, namely, with any combined parameters of sperm concentration <15 × 106/ml, and/or progressive motility <32%, and/or normal morphology <4%. The WES was performed for all 157 samples. Subsequent calling and filtering of variants were carried out according to the GATK Best Practices recommendations. On the genotyping stage, the samples were combined into four cohorts: three sets corresponded to three ethnic groups, and the fourth set contained all the 157 whole-exome samples. Association of the obtained polymorphisms with semen quality parameters was investigated using the χ2 test. To prioritize the obtained variants associated with pathozoospermia, their effects were determined using Ensembl Variant Effect Predictor. Moreover, polymorphisms located in genes expressed in the testis were revealed based on the genomic annotation. As a result, the nine potential SNP markers rs6971091, rs557806, rs610308, rs556052, rs1289658, rs278981, rs1129172, rs12268007, and rs17228441 were selected for subsequent verification on our previously collected population sample (about 1,500 males). The selected variants located in seven genes FAM71F1, PPP1R15A, TRIM45, PRAME, RBM47, WDFY4, and FSIP2 that are expressed in the testis and play an important role in cell proliferation, meiosis, and apoptosis.
Keywords: Buryats; Slavs; Yakuts; association analysis; normospermia; pathozoospermia; semen quality; whole-exome sequencing.
Copyright © 2021 Kolmykov, Vasiliev, Osadchuk, Kleschev and Osadchuk.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Mutations in the stromal antigen 3 (STAG3) gene cause male infertility due to meiotic arrest.Hum Reprod. 2019 Nov 1;34(11):2112-2119. doi: 10.1093/humrep/dez204. Hum Reprod. 2019. PMID: 31682730
-
Semen quality of young men from the general population in Baltic countries.Hum Reprod. 2017 Jun 1;32(6):1334-1340. doi: 10.1093/humrep/dex062. Hum Reprod. 2017. PMID: 28383690
-
An age-based sperm nomogram: the McGill reference guide.Hum Reprod. 2020 Oct 1;35(10):2213-2225. doi: 10.1093/humrep/deaa196. Hum Reprod. 2020. PMID: 32914183
-
Use of whole exome and genome sequencing in the identification of genetic causes of primary immunodeficiencies.Curr Opin Allergy Clin Immunol. 2012 Dec;12(6):623-8. doi: 10.1097/ACI.0b013e3283588ca6. Curr Opin Allergy Clin Immunol. 2012. PMID: 23095910 Review.
-
Tutorial: guidelines for quality filtering of whole-exome and whole-genome sequencing data for population-scale association analyses.Nat Protoc. 2025 Mar 28. doi: 10.1038/s41596-025-01169-1. Online ahead of print. Nat Protoc. 2025. PMID: 40155705 Review.
Cited by
-
Male Infertility: New Developments, Current Challenges, and Future Directions.World J Mens Health. 2024 Jul;42(3):502-517. doi: 10.5534/wjmh.230232. Epub 2024 Jan 2. World J Mens Health. 2024. PMID: 38164030 Free PMC article. Review.
-
Editorial: Bioinformatics of Genome Regulation, Volume II.Front Genet. 2021 Nov 8;12:795257. doi: 10.3389/fgene.2021.795257. eCollection 2021. Front Genet. 2021. PMID: 34819949 Free PMC article. No abstract available.
-
Integrative pigGTEx resource with GWAS reveals genetic mechanism underlying semen quality in boars.J Anim Sci Biotechnol. 2025 Jul 21;16(1):105. doi: 10.1186/s40104-025-01237-2. J Anim Sci Biotechnol. 2025. PMID: 40685336 Free PMC article.
References
-
- Amiri-Yekta A., Coutton C., Kherraf Z. E., Karaouzène T., Le Tanno P., Sanati M. H., et al. (2016). Whole-exome sequencing of familial cases of multiple morphological abnormalities of the sperm flagella (MMAF) reveals new DNAH1 mutations. Hum. Reprod. 31 2872–2880. 10.1093/humrep/dew262 - DOI - PubMed
-
- Cannarella R., Condorelli R. A., Paolacci S., Barbagallo F., Guerri G., Bertelli M., et al. (2021). Next-generation sequencing: toward an increase in the diagnostic yield in patients with apparently idiopathic spermatogenic failure. Asian J. Androl. 23 24–29. 10.4103/aja.aja_25_20 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

