Expression Profiling and Functional Analysis of Circular RNAs in Inner Mongolian Cashmere Goat Hair Follicles
- PMID: 34178035
- PMCID: PMC8226234
- DOI: 10.3389/fgene.2021.678825
Expression Profiling and Functional Analysis of Circular RNAs in Inner Mongolian Cashmere Goat Hair Follicles
Abstract
Background: Inner Mongolian cashmere goats have hair of excellent quality and high economic value, and the skin hair follicle traits of cashmere goats have a direct and important effect on cashmere yield and quality. Circular RNA has been studied in a variety of tissues and cells.
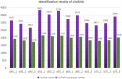
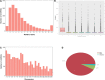
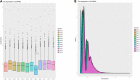
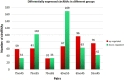
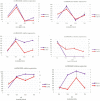
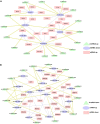
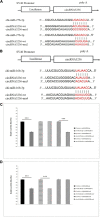
Result: In this study, high-throughput sequencing was used to obtain the expression profiles of circular RNA (circRNA) in the hair follicles of Inner Mongolian cashmere goats at different embryonic stages (45, 55, 65, and 75 days). A total of 21,784 circRNAs were identified. At the same time, the differentially expressed circRNA in the six comparison groups formed in the four stages were: d75vsd45, 59 upregulated and 33 downregulated DE circRNAs; d75vsd55, 61 upregulated and 102 downregulated DE circRNAs; d75vsd65, 32 upregulated and 33 downregulated DE circRNAs; d65vsd55, 67 upregulated and 169 downregulated DE circRNAs; d65vsd45, 96 upregulated and 63 downregulated DE circRNAs; and d55vsd45, 76 upregulated and 42 downregulated DE circRNAs. Six DE circRNA were randomly selected to verify the reliability of the sequencing results by quantitative RT-PCR. Subsequently, the circRNA corresponding host genes were analyzed by the Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. The results showed that the biological processes related to hair follicle growth and development enriched by GO mainly included hair follicle morphogenesis and cell development, and the signaling pathways related to hair follicle development included the Notch signaling pathway and NF-κB signaling pathway. We combined the DE circRNA of d75vsd45 with miRNA and mRNA databases (unpublished) to construct the regulatory network of circRNA-miRNA-mRNA, and formed a total of 102 pairs of circRNA-miRNA and 126 pairs of miRNA-mRNA interactions. The binding relationship of circRNA3236-chi-miR-27b-3p and circRNA3236-chi-miR-16b-3p was further verified by dual-luciferase reporter assays, and the results showed that circRNA3236 and chi-miR-27b-3p, and circRNA3236 and chi-miR-16b-3p have a targeted binding relationship.
Conclusion: To summarize, we established the expression profiling of circRNA in the fetal skin hair follicles of cashmere goats, and found that the host gene of circRNA may be involved in the development of hair follicles of cashmere goats. The regulatory network of circRNA-miRNA-mRNA was constructed and preliminarily verified using DE circRNAs.
Keywords: cashmere goat; circRNA; expression profile; functional analysis; hair follicles.
Copyright © 2021 Shang, Wang, Ma, Di, Wu, Hai, Rong, Pan, Liang, Wang, Wang, Liu, Zhao, Wang, Li and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Construction and functional analysis of ceRNA regulatory network related to the development of secondary hair follicles in Inner Mongolia cashmere goats.Front Vet Sci. 2022 Aug 25;9:959952. doi: 10.3389/fvets.2022.959952. eCollection 2022. Front Vet Sci. 2022. PMID: 36090177 Free PMC article.
-
Screening of microRNA and mRNA related to secondary hair follicle morphogenesis and development and functional analysis in cashmere goats.Funct Integr Genomics. 2022 Oct;22(5):835-848. doi: 10.1007/s10142-022-00842-y. Epub 2022 Apr 29. Funct Integr Genomics. 2022. PMID: 35488101 Free PMC article.
-
Inner Mongolian Cashmere Goat Secondary Follicle Development Regulation Research Based on mRNA-miRNA Co-analysis.Sci Rep. 2020 Mar 11;10(1):4519. doi: 10.1038/s41598-020-60351-5. Sci Rep. 2020. PMID: 32161290 Free PMC article.
-
RNA-Seq Revealed a Circular RNA-microRNA-mRNA Regulatory Network in Hantaan Virus Infection.Front Cell Infect Microbiol. 2020 Mar 13;10:97. doi: 10.3389/fcimb.2020.00097. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32232013 Free PMC article. Review.
-
Identification of circRNA biomarkers in osteosarcoma: An updated systematic review and meta-analysis.Noncoding RNA Res. 2024 Jan 11;9(2):341-349. doi: 10.1016/j.ncrna.2024.01.007. eCollection 2024 Jun. Noncoding RNA Res. 2024. PMID: 38505307 Free PMC article. Review.
Cited by
-
Multi-omics and AI-driven advances in miRNA-mediated hair follicle regulation in cashmere goats.Front Vet Sci. 2025 Jul 9;12:1635202. doi: 10.3389/fvets.2025.1635202. eCollection 2025. Front Vet Sci. 2025. PMID: 40703926 Free PMC article. Review.
-
An integrative analysis of the lncRNA-miRNA-mRNA competitive endogenous RNA network reveals potential mechanisms in the murine hair follicle cycle.Front Genet. 2022 Oct 25;13:931797. doi: 10.3389/fgene.2022.931797. eCollection 2022. Front Genet. 2022. PMID: 36386842 Free PMC article.
-
MicroRNA and circular RNA profiling in the deposited fat tissue of Sunite sheep.Front Vet Sci. 2022 Nov 4;9:954882. doi: 10.3389/fvets.2022.954882. eCollection 2022. Front Vet Sci. 2022. PMID: 36406061 Free PMC article.
-
Expression Profile Analysis to Identify Circular RNA Expression Signatures in the Prolificacy Trait of Yunshang Black Goat Pituitary in the Estrus Cycle.Front Genet. 2022 Jan 24;12:801357. doi: 10.3389/fgene.2021.801357. eCollection 2021. Front Genet. 2022. PMID: 35140742 Free PMC article.
-
Identification and Characterization of Circular RNAs (circRNAs) Using RNA-Seq in Two Breeds of Cashmere Goats.Genes (Basel). 2023 Jan 27;14(2):331. doi: 10.3390/genes14020331. Genes (Basel). 2023. PMID: 36833256 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

