Assembling Disease Networks From Causal Interaction Resources
- PMID: 34178043
- PMCID: PMC8226215
- DOI: 10.3389/fgene.2021.694468
Assembling Disease Networks From Causal Interaction Resources
Abstract
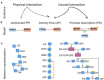
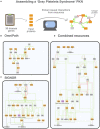
The development of high-throughput high-content technologies and the increased ease in their application in clinical settings has raised the expectation of an important impact of these technologies on diagnosis and personalized therapy. Patient genomic and expression profiles yield lists of genes that are mutated or whose expression is modulated in specific disease conditions. The challenge remains of extracting from these lists functional information that may help to shed light on the mechanisms that are perturbed in the disease, thus setting a rational framework that may help clinical decisions. Network approaches are playing an increasing role in the organization and interpretation of patients' data. Biological networks are generated by connecting genes or gene products according to experimental evidence that demonstrates their interactions. Till recently most approaches have relied on networks based on physical interactions between proteins. Such networks miss an important piece of information as they lack details on the functional consequences of the interactions. Over the past few years, a number of resources have started collecting causal information of the type protein A activates/inactivates protein B, in a structured format. This information may be represented as signed directed graphs where physiological and pathological signaling can be conveniently inspected. In this review we will (i) present and compare these resources and discuss the different scope in comparison with pathway resources; (ii) compare resources that explicitly capture causality in terms of data content and proteome coverage (iii) review how causal-graphs can be used to extract disease-specific Boolean networks.
Keywords: causal interactions; causality resources; logic modeling; network medicine; prior knowledge network.
Copyright © 2021 Cesareni, Sacco and Perfetto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Boué S., Talikka M., Westra J. W., Hayes W., Di Fabio A., Park J., et al. (2015). Causal biological network database: a comprehensive platform of causal biological network models focused on the pulmonary and vascular systems. Database J. Biol. Databases Curation 2015:bav030. 10.1093/database/bav030 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

