CXCL12 Retargeting of an Oncolytic Adenovirus Vector to the Chemokine CXCR4 and CXCR7 Receptors in Breast Cancer
- PMID: 34178415
- PMCID: PMC8225250
- DOI: 10.4236/jct.2021.126029
CXCL12 Retargeting of an Oncolytic Adenovirus Vector to the Chemokine CXCR4 and CXCR7 Receptors in Breast Cancer
Abstract
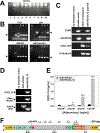
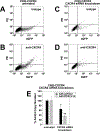
Breast cancer is the most frequently diagnosed cancer in women under 60, and the second most diagnosed cancer in women over 60. While significant progress has been made in developing targeted therapies for breast cancer, advanced breast cancer continues to have high mortality, with poor 5-year survival rates. Thus, current therapies are insufficient in treating advanced stages of breast cancer; new treatments are sorely needed to address the complexity of advanced-stage breast cancer. Oncolytic virotherapy has been explored as a therapeutic approach capable of systemic administration, targeting cancer cells, and sparing normal tissue. In particular, oncolytic adenoviruses have been exploited as viral vectors due to their ease of manipulation, production, and demonstrated clinical safety profile. In this study, we engineered an oncolytic adenovirus to target the chemokine receptors CXCR4 and CXCR7. The overexpression of CXCR4 and CXCR7 is implicated in the initiation, survival, progress, and metastasis of breast cancer. Both receptors bind to the ligand, CXCL12 (SDF-1), which has been identified to play a crucial role in the metastasis of breast cancer cells. This study incorporated a T4 fibritin protein fused to CXCL12 into the tail domain of an adenovirus fiber to retarget the vector to the CXCR4 and CXCR7 chemokine receptors. We showed that the modified virus targets and infects CXCR4- and CXCR7-overexpressing breast cancer cells more efficiently than a wild-type control vector. In addition, the substitution of the wild-type fiber and knob with the modified chimeric fiber did not interfere with oncolytic capability. Overall, the results of this study demonstrate the feasibility of retargeting adenovirus vectors to chemokine receptor-positive tumors.
Keywords: Adenovirus; Breast Cancer; CXCL12; CXCR4; CXCR7; Cancer; Chemokine; Oncolytic; Preclinical; Receptor; Virotherapy; Virus.
Conflict of interest statement
Conflicts of Interest The authors declare no conflict of interest. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures




References
Grants and funding
LinkOut - more resources
Full Text Sources
