Role of plasmonics in detection of deadliest viruses: a review
- PMID: 34178567
- PMCID: PMC8214556
- DOI: 10.1140/epjp/s13360-021-01657-9
Role of plasmonics in detection of deadliest viruses: a review
Abstract
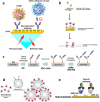
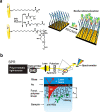
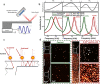
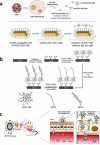
Viruses have threatened animal and human lives since a long time ago all over the world. Some of these tiny particles have caused disastrous pandemics that killed a large number of people with subsequent economic downturns. In addition, the quarantine situation itself encounters the challenges like the deficiency in the online educational system, psychiatric problems and poor international relations. Although viruses have a rather simple protein structure, they have structural heterogeneity with a high tendency to mutation that impedes their study. On top of the breadth of such worldwide worrying issues, there are profound scientific gaps, and several unanswered questions, like lack of vaccines or antivirals to combat these pathogens. Various detection techniques like the nucleic acid test, immunoassay, and microscopy have been developed; however, there is a tradeoff between their advantages and disadvantages like safety in sample collecting, invasiveness, sensitivity, response time, etc. One of the highly resolved techniques that can provide early-stage detection with fast experiment duration is plasmonics. This optical technique has the capability to detect viral proteins and genomes at the early stage via highly sensitive interaction between the biological target and the plasmonic chip. The efficiency of this technique could be proved using commercialized techniques like reverse transcription polymerase chain reaction (RT-PCR) and enzyme-linked immunosorbent assay (ELISA) techniques. In this study, we aim to review the role of plasmonic technique in the detection of 11 deadliest viruses besides 2 common genital viruses for the human being. This is a rapidly moving topic of research, and a review article that encompasses the current findings may be useful for guiding strategies to deal with the pandemics. By investigating the potential aspects of this technique, we hope that this study could open new avenues toward the application of point-of-care techniques for virus detection at early stage that may inhibit the progressively hygienic threats.
© The Author(s), under exclusive licence to Società Italiana di Fisica and Springer-Verlag GmbH Germany, part of Springer Nature 2021.
Figures






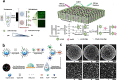
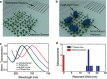

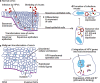

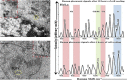

Similar articles
-
Diagnostic methods for detection & isolation of dengue viruses from vector mosquitoes.Indian J Med Res. 2006 May;123(5):615-28. Indian J Med Res. 2006. PMID: 16873905 Review.
-
Applications of Nanozymology in the Detection and Identification of Viral, Bacterial and Fungal Pathogens.Int J Mol Sci. 2022 Apr 22;23(9):4638. doi: 10.3390/ijms23094638. Int J Mol Sci. 2022. PMID: 35563029 Free PMC article. Review.
-
[Research progress on analysis of human papillomavirus by microchip capillary electrophoresis].Se Pu. 2020 Oct 8;38(10):1179-1188. doi: 10.3724/SP.J.1123.2020.05016. Se Pu. 2020. PMID: 34213114 Review. Chinese.
-
A one-step multiplex RT-PCR assay for simultaneous detection of four viruses that infect peach.Lett Appl Microbiol. 2013 Oct;57(4):350-5. doi: 10.1111/lam.12120. Epub 2013 Jul 8. Lett Appl Microbiol. 2013. PMID: 23777367
-
[Comparison study of a real-time reverse transcription polymerase chain reaction assay with an enzyme immunoassay and shell vial culture for influenza A and B virus detection in adult patients].Enferm Infecc Microbiol Clin. 2010 Feb;28(2):95-8. doi: 10.1016/j.eimc.2008.11.021. Epub 2009 May 23. Enferm Infecc Microbiol Clin. 2010. PMID: 19477042 Clinical Trial. Spanish.
Cited by
-
The Continuous Adaptive Challenge Played by Arboviruses: An In Silico Approach to Identify a Possible Interplay between Conserved Viral RNA Sequences and Host RNA Binding Proteins (RBPs).Int J Mol Sci. 2023 Jul 4;24(13):11051. doi: 10.3390/ijms241311051. Int J Mol Sci. 2023. PMID: 37446229 Free PMC article.
-
SPR-Based Kinetic Analysis of the Early Stages of Infection in Cells Infected with Human Coronavirus and Treated with Hydroxychloroquine.Biosensors (Basel). 2021 Jul 26;11(8):251. doi: 10.3390/bios11080251. Biosensors (Basel). 2021. PMID: 34436052 Free PMC article.
-
Smartphone-Based Multiplexed Biosensing Tools for Health Monitoring.Biosensors (Basel). 2022 Jul 29;12(8):583. doi: 10.3390/bios12080583. Biosensors (Basel). 2022. PMID: 36004979 Free PMC article. Review.
References
-
- Organization, W.H.; Available from: https://www.who.int/emergencies/diseases/novel-coronavirus-2019
-
- Worldometer. Available from: https://www.worldometers.info/coronavirus/
-
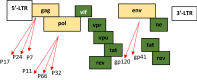
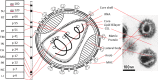
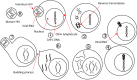
- B. Marintcheva, Chapter 1 - Introduction to Viral Structure, Diversity and Biology∗∗Parts of this chapter were originally published in Marintcheva B. A box of paradoxes: the fascinating world of viruses. Bridgew Rev 2013;32(2):25–8. http://vc.bridgew.edu/br_rev/vol32/iss2/8 and are reproduced here with the permission of the editor, in Harnessing the Power of Viruses, B. Marintcheva, Editor. 2018, Academic Press. p. 1–26
Publication types
LinkOut - more resources
Full Text Sources
