The lncRNA ADAMTS9-AS2 Regulates RPL22 to Modulate TNBC Progression via Controlling the TGF-β Signaling Pathway
- PMID: 34178640
- PMCID: PMC8219971
- DOI: 10.3389/fonc.2021.654472
The lncRNA ADAMTS9-AS2 Regulates RPL22 to Modulate TNBC Progression via Controlling the TGF-β Signaling Pathway
Abstract
Background: Long non-coding RNAs (lncRNAs) are key regulators of triple-negative breast cancer (TNBC) progression, but further work is needed to fully understand the functional relevance of these non-coding RNAs in this cancer type. Herein, we explored the functional role of the lncRNA ADAMTS9-AS2 in TNBC.
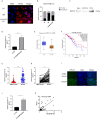
Methods: Next-generation sequencing was conducted to compare the expression of different lncRNAs in TNBC tumor and paracancerous tissues, after which ADAMTS9-AS2differential expression in these tumor tissues was evaluated via qPCR. The functional role of this lncRNA was assessed by overexpressing it in vitro and in vivo. FISH and PCR were used to assess the localization of ADAMTS9-AS2within cells. Downstream targets of ADAMTS9-AS2 signaling were identified via RNA pulldown assays and transcriptomic sequencing.
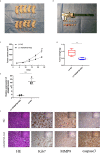
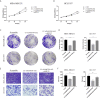
Results: The expression ofADAMTS9-AS2 was decreased in TNBC tumor samples (P < 0.05), with such downregulation being correlated with TNM stage, age, and tumor size. Overexpressing ADAMTS9-AS2 promoted the apoptotic death and cell cycle arrest of tumor cells in vitro and inhibited tumor growth in vivo. From a mechanistic perspective, ADAMTS9-AS2 was found to control the expression of RPL22 and to thereby modulate TGF-β signaling to control TNBC progression.
Conclusion: ADAMTS9-AS2 controls the expression of RPL22 and thereby regulates TNBC malignancy via the TGF-β signaling pathway.
Keywords: ADAMTS9-AS2; TGFb (transforming growth factor-beta); TNBC (Triple negative breast cancer); lncRNA; signaling pathway.
Copyright © 2021 Ni, Huang, Zhu, Xue, Jin, Zhang and Gu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
lncRNA ADAMTS9-AS2 Controls Human Mesenchymal Stem Cell Chondrogenic Differentiation and Functions as a ceRNA.Mol Ther Nucleic Acids. 2019 Dec 6;18:533-545. doi: 10.1016/j.omtn.2019.08.027. Epub 2019 Sep 18. Mol Ther Nucleic Acids. 2019. PMID: 31671346 Free PMC article.
-
The lncRNAs PART1 and ADAMTS9-AS2 act in an antithetic manner on AR signaling and induction of cellular senescence in prostate cancer cells.Int J Surg. 2025 May 1;111(5):3646-3664. doi: 10.1097/JS9.0000000000002334. Int J Surg. 2025. PMID: 40143747 Free PMC article.
-
LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway.Int J Biol Macromol. 2018 Dec;120(Pt B):1705-1713. doi: 10.1016/j.ijbiomac.2018.09.179. Epub 2018 Sep 27. Int J Biol Macromol. 2018. PMID: 30268751
-
ADAMTS9-AS2: A Functional Long Non-coding RNA in Tumorigenesis.Curr Pharm Des. 2021;27(23):2722-2727. doi: 10.2174/1381612827666210325105106. Curr Pharm Des. 2021. PMID: 33823762 Review.
-
A review on the role of ADAMTS9-AS2 in different disorders.Pathol Res Pract. 2023 Mar;243:154346. doi: 10.1016/j.prp.2023.154346. Epub 2023 Jan 27. Pathol Res Pract. 2023. PMID: 36746036 Review.
Cited by
-
Exploring the clinical potential of circulating LncRNAs in breast cancer: insights into primary signaling pathways and therapeutic interventions.Funct Integr Genomics. 2024 Nov 7;24(6):209. doi: 10.1007/s10142-024-01476-y. Funct Integr Genomics. 2024. PMID: 39508907 Review.
-
Roles of TGF‑β signalling pathway‑related lncRNAs in cancer (Review).Oncol Lett. 2023 Feb 2;25(3):107. doi: 10.3892/ol.2023.13693. eCollection 2023 Mar. Oncol Lett. 2023. PMID: 36817052 Free PMC article. Review.
-
Identification of long noncoding RNAs downregulated specifically in ovarian high-grade serous carcinoma.Reprod Med Biol. 2024 Apr 3;23(1):e12572. doi: 10.1002/rmb2.12572. eCollection 2024 Jan-Dec. Reprod Med Biol. 2024. PMID: 38571514 Free PMC article.
-
The long non-coding RNAs (lncRNA) in the pathogenesis of gastric cancer cells: molecular mechanisms and involvement miRNAs.Mol Biol Rep. 2024 May 5;51(1):615. doi: 10.1007/s11033-024-09546-x. Mol Biol Rep. 2024. PMID: 38704760 Review.
-
Long non-coding RNAs as the critical regulators of PI3K/AKT, TGF-β, and MAPK signaling pathways during breast tumor progression.J Transl Med. 2023 Aug 18;21(1):556. doi: 10.1186/s12967-023-04434-7. J Transl Med. 2023. PMID: 37596669 Free PMC article. Review.
References
-
- Beek MA, Gobardhan PD, Klompenhouwer EG, Menke-Pluijmers MB, Steenvoorde P, Merkus JW, et al. . A Patient- and Assessor-Blinded Randomized Controlled Trial of Axillary Reverse Mapping (ARM) in Patients With Early Breast Cancer. Eur J Surg Oncol (2020) 46(1):59–64. 10.1016/j.ejso.2019.08.003 - DOI - PubMed
LinkOut - more resources
Full Text Sources

