Leveraging 16S rRNA Microbiome Sequencing Data to Identify Bacterial Signatures for Irritable Bowel Syndrome
- PMID: 34178718
- PMCID: PMC8231010
- DOI: 10.3389/fcimb.2021.645951
Leveraging 16S rRNA Microbiome Sequencing Data to Identify Bacterial Signatures for Irritable Bowel Syndrome
Abstract
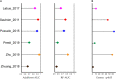
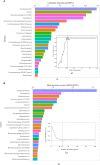
Irritable bowel syndrome (IBS) is a chronic gastrointestinal disorder characterized by abdominal pain or discomfort. Previous studies have illustrated that the gut microbiota might play a critical role in IBS, but the conclusions of these studies, based on various methods, were almost impossible to compare, and reproducible microorganism signatures were still in question. To cope with this problem, previously published 16S rRNA gene sequencing data from 439 fecal samples, including 253 IBS samples and 186 control samples, were collected and processed with a uniform bioinformatic pipeline. Although we found no significant differences in community structures between IBS and healthy controls at the amplicon sequence variants (ASV) level, machine learning (ML) approaches enabled us to discriminate IBS from healthy controls at genus level. Linear discriminant analysis effect size (LEfSe) analysis was subsequently used to seek out 97 biomarkers across all studies. Then, we quantified the standardized mean difference (SMDs) for all significant genera identified by LEfSe and ML approaches. Pooled results showed that the SMDs of nine genera had statistical significance, in which the abundance of Lachnoclostridium, Dorea, Erysipelatoclostridium, Prevotella 9, and Clostridium sensu stricto 1 in IBS were higher, while the dominant abundance genera of healthy controls were Ruminococcaceae UCG-005, Holdemanella, Coprococcus 2, and Eubacterium coprostanoligenes group. In summary, based on six published studies, this study identified nine new microbiome biomarkers of IBS, which might be a basis for understanding the key gut microbes associated with IBS, and could be used as potential targets for microbiome-based diagnostics and therapeutics.
Keywords: 16S rRNA; biomarkers; gut microbiome; irritable bowel syndrome; machine learning algorithm.
Copyright © 2021 Liu, Li, Yang, Zhang, Wang, Jia, Xiang, Wang, Miao, Zhang, Wang, Wang, Song, Sun, Chai and Tian.
Conflict of interest statement
Authors LW, YW, JS and YS were employed by company Tianjin Zhongxin Pharmaceutical Group Co., Ltd. The reamining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

