Candida albicans SET3 Plays a Role in Early Biofilm Formation, Interaction With Pseudomonas aeruginosa and Virulence in Caenorhabditis elegans
- PMID: 34178723
- PMCID: PMC8223063
- DOI: 10.3389/fcimb.2021.680732
Candida albicans SET3 Plays a Role in Early Biofilm Formation, Interaction With Pseudomonas aeruginosa and Virulence in Caenorhabditis elegans
Abstract
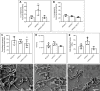
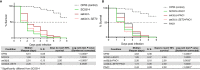
The yeast Candida albicans exhibits multiple morphologies dependent on environmental cues. Candida albicans biofilms are frequently polymicrobial, enabling interspecies interaction through proximity and contact. The interaction between C. albicans and the bacterium, Pseudomonas aeruginosa, is antagonistic in vitro, with P. aeruginosa repressing the yeast-to-hyphal switch in C. albicans. Previous transcriptional analysis of C. albicans in polymicrobial biofilms with P. aeruginosa revealed upregulation of genes involved in regulation of morphology and biofilm formation, including SET3, a component of the Set3/Hos2 histone deacetylase complex (Set3C). This prompted the question regarding the involvement of SET3 in the interaction between C. albicans and P. aeruginosa, both in vitro and in vivo. We found that SET3 may influence early biofilm formation by C. albicans and the interaction between C. albicans and P. aeruginosa. In addition, although deletion of SET3 did not alter the morphology of C. albicans in the presence of P. aeruginosa, it did cause a reduction in virulence in a Caenorhabditis elegans infection model, even in the presence of P. aeruginosa.
Keywords: Caenorhabditis elegans; Candida albicans; Pseudomonas aeruginosa; SET3; biofilm.
Copyright © 2021 Fourie, Albertyn, Sebolai, Gcilitshana and Pohl.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Recent Advances and Opportunities in the Study of Candida albicans Polymicrobial Biofilms.Front Cell Infect Microbiol. 2022 Feb 18;12:836379. doi: 10.3389/fcimb.2022.836379. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35252039 Free PMC article. Review.
-
Role of the high-affinity reductive iron acquisition pathway of Candida albicans in prostaglandin E2 production, virulence, and interaction with Pseudomonas aeruginosa.Med Mycol. 2021 Sep 3;59(9):869-881. doi: 10.1093/mmy/myab015. Med Mycol. 2021. PMID: 33862618
-
Transcriptional response of Candida albicans to Pseudomonas aeruginosa in a polymicrobial biofilm.G3 (Bethesda). 2021 Apr 15;11(4):jkab042. doi: 10.1093/g3journal/jkab042. G3 (Bethesda). 2021. PMID: 33580263 Free PMC article.
-
Both Pseudomonas aeruginosa and Candida albicans Accumulate Greater Biomass in Dual-Species Biofilms under Flow.mSphere. 2021 Jun 30;6(3):e0041621. doi: 10.1128/mSphere.00416-21. Epub 2021 Jun 23. mSphere. 2021. PMID: 34160236 Free PMC article.
-
Candida albicans and Pseudomonas aeruginosa interactions: more than an opportunistic criminal association?Med Mal Infect. 2013 Apr;43(4):146-51. doi: 10.1016/j.medmal.2013.02.005. Epub 2013 Apr 23. Med Mal Infect. 2013. PMID: 23622953 Review.
Cited by
-
Recent Advances and Opportunities in the Study of Candida albicans Polymicrobial Biofilms.Front Cell Infect Microbiol. 2022 Feb 18;12:836379. doi: 10.3389/fcimb.2022.836379. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35252039 Free PMC article. Review.
-
Current and prospective therapeutic strategies: tackling Candida albicans and Streptococcus mutans cross-kingdom biofilm.Front Cell Infect Microbiol. 2023 May 11;13:1106231. doi: 10.3389/fcimb.2023.1106231. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37249973 Free PMC article. Review.
-
Polymicrobial Infections and Biofilms: Clinical Significance and Eradication Strategies.Antibiotics (Basel). 2022 Dec 1;11(12):1731. doi: 10.3390/antibiotics11121731. Antibiotics (Basel). 2022. PMID: 36551388 Free PMC article. Review.
-
Properties and biotechnological applications of microbial deacetylase.Appl Microbiol Biotechnol. 2023 Aug;107(15):4697-4716. doi: 10.1007/s00253-023-12613-1. Epub 2023 Jun 16. Appl Microbiol Biotechnol. 2023. PMID: 37326683 Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

