ITGA2 promotes expression of ACLY and CCND1 in enhancing breast cancer stemness and metastasis
- PMID: 34179312
- PMCID: PMC8209312
- DOI: 10.1016/j.gendis.2020.01.015
ITGA2 promotes expression of ACLY and CCND1 in enhancing breast cancer stemness and metastasis
Abstract
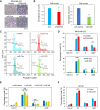
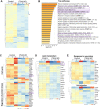
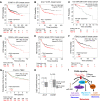
Cancer metastasis is largely incurable and accounts for 90% of breast cancer deaths, especially for the aggressive basal-like or triple negative breast cancer (TNBC). Combining patient database analyses and functional studies, we examined the association of integrin family members with clinical outcomes as well as their connection with previously identified microRNA regulators of metastasis, such as miR-206 that inhibits stemness and metastasis of TNBC. Here we report that the integrin receptor CD49b-encoding ITGA2, a direct target of miR-206, promotes breast cancer stemness and metastasis. ITGA2 knockdown suppressed self-renewal related mammosphere formation and pluripotency marker expression, inhibited cell cycling, compromised migration and invasion, and therefore decreased lung metastasis of breast cancer. ITGA2 overexpression reversed miR-206-caused cell cycle arrest in G1. RNA sequencing analyses revealed that ITGA2 knockdown inhibits genes related to cell cycle regulation and lipid metabolism, including CCND1 and ACLY as representative targets, respectively. Knockdown of CCND1 or ACLY inhibits mammosphere formation of breast cancer cells. Overexpression of CCND1 rescues the phenotype of ITGA2 knockdown-induced cell cycle arrest. ACLY-encoded ATP citrate lyase is essential to maintain cellular acetyl-CoA levels. CCND1 knockdown further mimics ITGA2 knockdown in abolishing lung colonization of breast cancer cells. We identified that the low levels of miR-206 as well as high expression levels of ITGA2, ACLY and CCND1 are associated with an unfavorable relapse-free survival of the patients with estrogen receptor-negative or high grade breast cancer, especially basal-like or TNBC, possibly serving as potential biomarkers of cancer stemness and therapeutic targets of breast cancer metastasis.
Keywords: ACLY; Breast cancer; CCND1; CD49b; ITGA2; Integrins; Metastasis; Stemness.
© 2020 The Authors.
Figures




References
-
- Cancer IAfRo . The Glocal Cancer Observatory; 2018. All cancers. [online]
-
- Howlader N.N.A., Noone A.M., Krapcho M. National Cancer Institute; 2016. SEER Cancer Statistics Review.
-
- Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA A Cancer J Clin. 2018;68(1):7–30. - PubMed
-
- Lapidot T., Sirard C., Vormoor J. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

