Cardiac troponin I R193H mutant interacts with HDAC1 to repress phosphodiesterase 4D expression in cardiomyocytes
- PMID: 34179318
- PMCID: PMC8209310
- DOI: 10.1016/j.gendis.2020.01.004
Cardiac troponin I R193H mutant interacts with HDAC1 to repress phosphodiesterase 4D expression in cardiomyocytes
Abstract
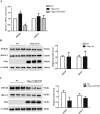
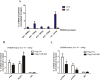
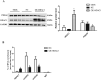
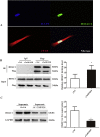
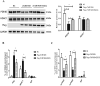
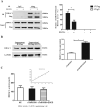
Cardiac Troponin I (cTnI) is a subunit of the thin filament involved in regulation of heart contraction. Mutated cTnI accounts for most genetic mutations associated with restrictive cardiomyopathy (RCM). We previously found phosphodiesterase 4D (PDE4D) decreased in RCM mice with cTnIR193H mutation and the mutant cTnI might be involved in PDE4D reduction. This study aims to elucidate a novel role of cTnIR193H mutant as a gene regulator. Overexpression of cTnIR193H mutant in cardiomyocytes showed decrease in PDED4D protein expression, while the enrichment of histone deacetylase 1 (HDAC1) was increased along with decreases in acetylated lysine 4 (acH3K4) and 9 (acH3K9) levels in the PDE4D promoter. HDAC1 overexpression could also downregulate PDE4D via reducing acH3K4 and acH3K9 levels. Co-IP assays showed that cTnIR193H mutant owed increased binding ability to HDAC1 compared with wild type cTnI. EGCG as a HDAC1 inhibitor could diminish the strength of cTnIR193H-HDAC1 interactions and alleviate the reduction in PDE4D expression. Together, our data indicated that cTnIR193H mutant could repress PDE4D expression in cardiomyocytes through HDAC1 associated histone deacetylation modification. Unlike the typical function of cTnI in cytoplasm, our study suggested a novel role of cTnI mutants in nuclei in regulating gene expression.
Keywords: EGCG; HDAC1; Histone modifications; PDE4D reduction; cTnIR93H.
© 2020 Chongqing Medical University. Production and hosting by Elsevier B.V.
Figures

Similar articles
-
Epigenetic regulation of phosphodiesterase 4d in restrictive cardiomyopathy mice with cTnI mutations.Sci China Life Sci. 2020 Apr;63(4):563-570. doi: 10.1007/s11427-018-9463-9. Epub 2019 Mar 12. Sci China Life Sci. 2020. PMID: 30900165
-
Epigallocatechin gallate reverses cTnI-low expression-induced age-related heart diastolic dysfunction through histone acetylation modification.J Cell Mol Med. 2017 Oct;21(10):2481-2490. doi: 10.1111/jcmm.13169. Epub 2017 Apr 6. J Cell Mol Med. 2017. PMID: 28382690 Free PMC article.
-
Cardiac Troponin I R193H Mutation Is Associated with Mitochondrial Damage in Cardiomyocytes.DNA Cell Biol. 2021 Feb;40(2):184-191. doi: 10.1089/dna.2020.5828. Epub 2021 Jan 18. DNA Cell Biol. 2021. PMID: 33465007
-
Restrictive Cardiomyopathy Caused by Troponin Mutations: Application of Disease Animal Models in Translational Studies.Front Physiol. 2016 Dec 19;7:629. doi: 10.3389/fphys.2016.00629. eCollection 2016. Front Physiol. 2016. PMID: 28066262 Free PMC article. Review.
-
Biology of the troponin complex in cardiac myocytes.Prog Cardiovasc Dis. 2004 Nov-Dec;47(3):159-76. doi: 10.1016/j.pcad.2004.07.003. Prog Cardiovasc Dis. 2004. PMID: 15736582 Review.
Cited by
-
Unlocking cardiac health: exploring the role of class I HDACs in cardiovascular diseases.Mol Cell Biochem. 2025 Jul 14. doi: 10.1007/s11010-025-05353-5. Online ahead of print. Mol Cell Biochem. 2025. PMID: 40660005 Review.
-
The effect of long-term administration of green tea catechins on aging-related cardiac diastolic dysfunction and decline of troponin I.Genes Dis. 2024 Apr 3;12(2):101284. doi: 10.1016/j.gendis.2024.101284. eCollection 2025 Mar. Genes Dis. 2024. PMID: 39759124 Free PMC article.
-
Epigallocatechin-3-gallate restores mitochondrial homeostasis impairment by inhibiting HDAC1-mediated NRF1 histone deacetylation in cardiac hypertrophy.Mol Cell Biochem. 2024 Apr;479(4):963-973. doi: 10.1007/s11010-023-04768-2. Epub 2023 Jun 2. Mol Cell Biochem. 2024. PMID: 37266748
-
Epigenetic Mechanisms in Heart Diseases.Rev Cardiovasc Med. 2025 Jul 30;26(7):38696. doi: 10.31083/RCM38696. eCollection 2025 Jul. Rev Cardiovasc Med. 2025. PMID: 40776938 Free PMC article. Review.
-
The Emerging Role of Epigenetics in Therapeutic Targeting of Cardiomyopathies.Int J Mol Sci. 2021 Aug 13;22(16):8721. doi: 10.3390/ijms22168721. Int J Mol Sci. 2021. PMID: 34445422 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

