A deep learning approach to predict blood-brain barrier permeability
- PMID: 34179448
- PMCID: PMC8205267
- DOI: 10.7717/peerj-cs.515
A deep learning approach to predict blood-brain barrier permeability
Abstract
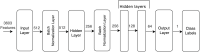
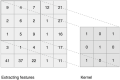
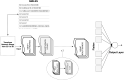
The blood-brain barrier plays a crucial role in regulating the passage of 98% of the compounds that enter the central nervous system (CNS). Compounds with high permeability must be identified to enable the synthesis of brain medications for the treatment of various brain diseases, such as Parkinson's, Alzheimer's, and brain tumors. Throughout the years, several models have been developed to solve this problem and have achieved acceptable accuracy scores in predicting compounds that penetrate the blood-brain barrier. However, predicting compounds with "low" permeability has been a challenging task. In this study, we present a deep learning (DL) classification model to predict blood-brain barrier permeability. The proposed model addresses the fundamental issues presented in former models: high dimensionality, class imbalances, and low specificity scores. We address these issues to enhance the high-dimensional, imbalanced dataset before developing the classification model: the imbalanced dataset is addressed using oversampling techniques and the high dimensionality using a non-linear dimensionality reduction technique known as kernel principal component analysis (KPCA). This technique transforms the high-dimensional dataset into a low-dimensional Euclidean space while retaining invaluable information. For the classification task, we developed an enhanced feed-forward deep learning model and a convolutional neural network model. In terms of specificity scores (i.e., predicting compounds with low permeability), the results obtained by the enhanced feed-forward deep learning model outperformed those obtained by other models in the literature that were developed using the same technique. In addition, the proposed convolutional neural network model surpassed models used in other studies in multiple accuracy measures, including overall accuracy and specificity. The proposed approach solves the problem inevitably faced with obtaining low specificity resulting in high false positive rate.
Keywords: Blood Brain Barrier (BBB) permeability; Chemoinformatics; Convolutional Neural Network (CNN); Quantitative Structure-Activity Relationships (QSAR).
©2021 Alsenan et al.
Conflict of interest statement
The authors declare they have no competing interests.
Figures




References
-
- Alsenan S, Al-Turaiki I, Hafez A. Autoencoder-based dimensionality reduction for QSAR modeling. 3rd international conference on computer applications and information security riyadh, Saudi Arabia, March 19-21, 2020; 2020. p. 65.
-
- Alvascience Srl alvaDesc. 2019. https://www.alvascience.com. [16 October 2019]. https://www.alvascience.com
-
- Arlot S, Celisse A. A survey of cross-validation procedures for model selection. Statistics Surveys. 2010;4:40–79.
LinkOut - more resources
Full Text Sources
