Multi-dimensional and longitudinal systems profiling reveals predictive pattern of severe COVID-19
- PMID: 34179733
- PMCID: PMC8213514
- DOI: 10.1016/j.isci.2021.102752
Multi-dimensional and longitudinal systems profiling reveals predictive pattern of severe COVID-19
Abstract
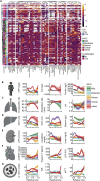
COVID-19 is a respiratory tract infection that can affect multiple organ systems. Predicting the severity and clinical outcome of individual patients is a major unmet clinical need that remains challenging due to intra- and inter-patient variability. Here, we longitudinally profiled and integrated more than 150 clinical, laboratory, and immunological parameters of 173 patients with mild to fatal COVID-19. Using systems biology, we detected progressive dysregulation of multiple parameters indicative of organ damage that correlated with disease severity, particularly affecting kidneys, hepatobiliary system, and immune landscape. By performing unsupervised clustering and trajectory analysis, we identified T and B cell depletion as early indicators of a complicated disease course. In addition, markers of hepatobiliary damage emerged as robust predictor of lethal outcome in critically ill patients. This allowed us to propose a novel clinical COVID-19 SeveriTy (COST) score that distinguishes complicated disease trajectories and predicts lethal outcome in critically ill patients.
Keywords: Immunology; Virology; systems biology.
© 2021 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- Becht E., McInnes L., Healy J., Dutertre C.-A., Kwok I.W.H., Ng L.G., Ginhoux F., Newell E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019;37:38–44. - PubMed
LinkOut - more resources
Full Text Sources

