Resident and circulating memory T cells persist for years in melanoma patients with durable responses to immunotherapy
- PMID: 34179824
- PMCID: PMC8223731
- DOI: 10.1038/s43018-021-00180-1
Resident and circulating memory T cells persist for years in melanoma patients with durable responses to immunotherapy
Abstract
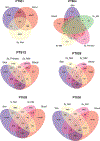
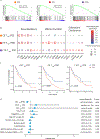
While T-cell responses to cancer immunotherapy have been avidly studied, long-lived memory has been poorly characterized. In a cohort of metastatic melanoma survivors with exceptional responses to immunotherapy, we probed memory CD8+ T-cell responses across tissues, and across several years. Single-cell RNA sequencing revealed three subsets of resident memory T (TRM) cells shared between tumors and distant vitiligo-affected skin. Paired T-cell receptor sequencing further identified clonotypes in tumors that co-existed as TRM in skin and as effector memory T (TEM) cells in blood. Clonotypes that dispersed throughout tumor, skin, and blood preferentially expressed a IFNG / TNF-high signature, which had a strong prognostic value for melanoma patients. Remarkably, clonotypes from tumors were found in patient skin and blood up to nine years later, with skin maintaining the most focused tumor-associated clonal repertoire. These studies reveal that cancer survivors can maintain durable memory as functional, broadly-distributed TRM and TEM compartments.
Conflict of interest statement
Competing Interests Statement All authors declare no competing interests.
Figures












Comment in
-
Melanoma-reactive T cells take up residence.Nat Cancer. 2021 Mar;2(3):253-255. doi: 10.1038/s43018-021-00189-6. Nat Cancer. 2021. PMID: 35121962 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

