Quantum processor-inspired machine learning in the biomedical sciences
- PMID: 34179840
- PMCID: PMC8212142
- DOI: 10.1016/j.patter.2021.100246
Quantum processor-inspired machine learning in the biomedical sciences
Abstract
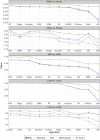
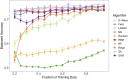
Recent advances in high-throughput genomic technologies coupled with exponential increases in computer processing and memory have allowed us to interrogate the complex molecular underpinnings of human disease from a genome-wide perspective. While the deluge of genomic information is expected to increase, a bottleneck in conventional high-performance computing is rapidly approaching. Inspired by recent advances in physical quantum processors, we evaluated several unconventional machine-learning (ML) strategies on actual human tumor data, namely "Ising-type" methods, whose objective function is formulated identical to simulated annealing and quantum annealing. We show the efficacy of multiple Ising-type ML algorithms for classification of multi-omics human cancer data from The Cancer Genome Atlas, comparing these classifiers to a variety of standard ML methods. Our results indicate that Ising-type ML offers superior classification performance with smaller training datasets, thus providing compelling empirical evidence for the potential future application of unconventional computing approaches in the biomedical sciences.
Keywords: The Cancer Genome Atlas; cancer genomics; machine learning.
© 2021 The Authors.
Conflict of interest statement
O.E.G., S.G., J.R.G., N.C., S.R.B., and T.W.C. were employed by Genuity Science during the research project. R.Y.L. was the recipient of a research grant from Genuity Science during the research project. The work of D.A.L. is based upon work (partially) supported by the Office of the Director of National Intelligence (ODNI), Intelligence Advanced Research Projects Activity (IARPA), via a US Army Research Office contract. The views and conclusions contained herein are those of the authors and should not be interpreted as necessarily representing the official policies or endorsements, either expressed or implied, of the ODNI, IARPA, or US government. The US government is authorized to reproduce and distribute reprints for governmental purposes notwithstanding any copyright annotation thereon. The authors declare no other competing interests.
Figures


References
-
- Golub T.R., Slonim D.K., Tamayo P., Huard C., Gaasenbeek M., Mesirov J.P., Coller H., Loh M.L., Downing J.R., Caligiuri M.A. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. - PubMed
-
- Nevins J.R., Potti A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat. Rev. Genet. 2007;8:601–609. - PubMed
-
- Uhlen M., Zhang C., Lee S., Sjöstedt E., Fagerberg L., Bidkhori G., Benfeitas R., Arif M., Liu Z., Edfors F. A pathology atlas of the human cancer transcriptome. Science. 2017;357:eaan2507. - PubMed
LinkOut - more resources
Full Text Sources

