Causal interactions from proteomic profiles: Molecular data meet pathway knowledge
- PMID: 34179843
- PMCID: PMC8212145
- DOI: 10.1016/j.patter.2021.100257
Causal interactions from proteomic profiles: Molecular data meet pathway knowledge
Abstract
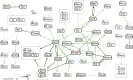
We present a computational method to infer causal mechanisms in cell biology by analyzing changes in high-throughput proteomic profiles on the background of prior knowledge captured in biochemical reaction knowledge bases. The method mimics a biologist's traditional approach of explaining changes in data using prior knowledge but does this at the scale of hundreds of thousands of reactions. This is a specific example of how to automate scientific reasoning processes and illustrates the power of mapping from experimental data to prior knowledge via logic programming. The identified mechanisms can explain how experimental and physiological perturbations, propagating in a network of reactions, affect cellular responses and their phenotypic consequences. Causal pathway analysis is a powerful and flexible discovery tool for a wide range of cellular profiling data types and biological questions. The automated causation inference tool, as well as the source code, are freely available at http://causalpath.org.
Keywords: cancer; causal pathway analysis; proteomics.
© 2021 The Authors.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Grants and funding
LinkOut - more resources
Full Text Sources

