Reprogramming of DNA methylation is linked to successful human preimplantation development
- PMID: 34179999
- PMCID: PMC8460514
- DOI: 10.1007/s00418-021-02008-6
Reprogramming of DNA methylation is linked to successful human preimplantation development
Abstract
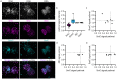
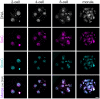
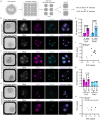
Human preimplantation development is characterized by low developmental rates that are poorly understood. Early mammalian embryogenesis is characterized by a major phase of epigenetic reprogramming, which involves global DNA methylation changes and activity of TET enzymes; the importance of DNA methylation reprogramming for successful human preimplantation development has not been investigated. Here, we analyzed early human embryos for dynamic changes in 5-methylcytosine and its oxidized derivatives generated by TET enzymes. We observed that 5-methylcytosine and 5-hydroxymethylcytosine show similar, albeit less pronounced, asymmetry between the parental pronuclei of human zygotes relative to mouse zygotes. Notably, we detected low levels of 5-formylcytosine and 5-carboxylcytosine, with no apparent difference in maternal or paternal pronuclei of human zygotes. Analysis of later human preimplantation stages revealed a mosaic pattern of DNA 5C modifications similar to those of the mouse and other mammals. Strikingly, using noninvasive time-lapse imaging and well-defined cell cycle parameters, we analyzed normally and abnormally developing human four-cell embryos for global reprogramming of DNA methylation and detected lower 5-methylcytosine and 5-hydroxymethylcytosine levels in normal embryos compared to abnormal embryos. In conclusion, our results suggest that DNA methylation reprogramming is conserved in humans, with human-specific dynamics and extent. Furthermore, abnormalities in the four-cell-specific DNA methylome in early human embryogenesis are associated with abnormal development, highlighting an essential role of epigenetic reprogramming for successful human embryogenesis. Further research should identify the underlying genomic regions and cause of abnormal DNA methylation reprogramming in early human embryos.
Keywords: Active DNA demethylation; DNA methylation reprogramming; Human preimplantation development; Successful human embryogenesis.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, Encheva V, Kudo NR, Koseki H, Sasaki H, Hajkova P. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol. 2016;18:225–233. doi: 10.1038/ncb3296. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

