The Urinary Microbiome in Postmenopausal Women with Recurrent Urinary Tract Infections
- PMID: 34181466
- PMCID: PMC8497440
- DOI: 10.1097/JU.0000000000001940
The Urinary Microbiome in Postmenopausal Women with Recurrent Urinary Tract Infections
Abstract
Purpose: The etiology of postmenopausal recurrent urinary tract infection (UTI) is not completely known, but the urinary microbiome is thought to be implicated. We compared the urinary microbiome in menopausal women with recurrent UTIs to age-matched controls, both in the absence of acute infection.
Materials and methods: This is a cross-sectional analysis of baseline data from 64 women enrolled in a longitudinal cohort study. All women were using topically applied vaginal estrogen. Women >55 years of age from the following groups were enrolled: 1) recurrent UTIs on daily antibiotic prophylaxis, 2) recurrent UTIs not on antibiotic prophylaxis and 3) age-matched controls without recurrent UTIs. Catheterized urine samples were collected at least 4 weeks after last treatment for UTI and at least 6 weeks after initiation of vaginal estrogen. Samples were evaluated using expanded quantitative urine culture (EQUC) and 16S rRNA gene sequencing.
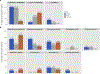
Results: With EQUC, there were no significant differences in median numbers of microbial species isolated among groups (p=0.96), even when considering Lactobacilli (p=0.72). However, there were trends toward different Lactobacillus species between groups. With 16S rRNA sequencing, the majority of urine samples contained Lactobacillaceae, with nonsignificant trends in relative abundance among groups. Using a Bayesian analysis, we identified significant differences in anaerobic taxa associated with phenotypic groups. Most of these differences centered on Bacteroidales and the family Prevotellaceae, although differences were also noted in Actinobacteria and certain genera of Clostridiales.
Conclusions: Associations between anaerobes within the urinary microbiome and postmenopausal recurrent UTI warrants further investigation.
Keywords: microbiota; postmenopause; urinary tract infections.
Figures




References
-
- Fouts DE, Pieper R, Szpakowski S, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Transl Med. August 28 2012;10:174. doi:10.1186/1479-5876-10-174 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

