SLIT2/ROBO signaling in tumor-associated microglia and macrophages drives glioblastoma immunosuppression and vascular dysmorphia
- PMID: 34181595
- PMCID: PMC8363292
- DOI: 10.1172/JCI141083
SLIT2/ROBO signaling in tumor-associated microglia and macrophages drives glioblastoma immunosuppression and vascular dysmorphia
Abstract
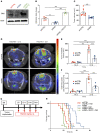
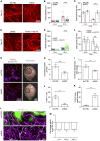
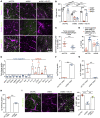
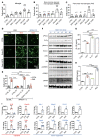
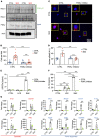
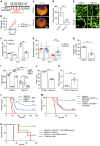
SLIT2 is a secreted polypeptide that guides migration of cells expressing Roundabout 1 and 2 (ROBO1 and ROBO2) receptors. Herein, we investigated SLIT2/ROBO signaling effects in gliomas. In patients with glioblastoma (GBM), SLIT2 expression increased with malignant progression and correlated with poor survival and immunosuppression. Knockdown of SLIT2 in mouse glioma cells and patient-derived GBM xenografts reduced tumor growth and rendered tumors sensitive to immunotherapy. Tumor cell SLIT2 knockdown inhibited macrophage invasion and promoted a cytotoxic gene expression profile, which improved tumor vessel function and enhanced efficacy of chemotherapy and immunotherapy. Mechanistically, SLIT2 promoted microglia/macrophage chemotaxis and tumor-supportive polarization via ROBO1- and ROBO2-mediated PI3K-γ activation. Macrophage Robo1 and Robo2 deletion and systemic SLIT2 trap delivery mimicked SLIT2 knockdown effects on tumor growth and the tumor microenvironment (TME), revealing SLIT2 signaling through macrophage ROBOs as a potentially novel regulator of the GBM microenvironment and immunotherapeutic target for brain tumors.
Keywords: Brain cancer; Immunotherapy; Macrophages; Oncology; Vascular Biology.
Conflict of interest statement
Figures