Diverse Humoral Immune Responses in Younger and Older Adult COVID-19 Patients
- PMID: 34182775
- PMCID: PMC8262923
- DOI: 10.1128/mBio.01229-21
Diverse Humoral Immune Responses in Younger and Older Adult COVID-19 Patients
Abstract
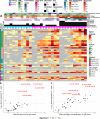
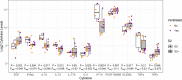
We sought to discover links between antibody responses to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and patient clinical variables, cytokine profiles, and antibodies to endemic coronaviruses. Serum samples from 30 patients of younger (26 to 39 years) and older (69 to 83 years) age groups and with varying clinical severities ranging from outpatient to mechanically ventilated were collected and used to probe a novel multi-coronavirus protein microarray. This microarray contained variable-length overlapping fragments of SARS-CoV-2 spike (S), envelope (E), membrane (M), nucleocapsid (N), and open reading frame (ORF) proteins created through in vitro transcription and translation (IVTT). The array also contained SARS-CoV, Middle East respiratory syndrome coronavirus (MERS-CoV), human coronavirus OC43 (HCoV-OC43), and HCoV-NL63 proteins. IgG antibody responses to specific epitopes within the S1 protein region spanning amino acids (aa) 500 to 650 and within the N protein region spanning aa 201 to 300 were found to be significantly higher in older patients and further significantly elevated in those older patients who were ventilated. Additionally, there was a noticeable overlap between antigenic regions and known mutation locations in selected emerging SARS-CoV-2 variants of current clinical consequence (B.1.1.7, B1.351, P.1, CAL20.C, and B.1.526). Moreover, the older age group displayed more consistent correlations of antibody reactivity with systemic cytokine and chemokine responses than the younger adult group. A subset of patients, however, had little or no response to SARS-CoV-2 antigens and disproportionately severe clinical outcomes. Further characterization of these slow-low-responding individuals with cytokine analysis revealed significantly higher interleukin-10 (IL-10), IL-15, and interferon gamma-induced protein 10 (IP-10) levels and lower epidermal growth factor (EGF) and soluble CD40 ligand (sCD40L) levels than those of seroreactive patients in the cohort. IMPORTANCE As numerous viral variants continue to emerge in the coronavirus disease 2019 (COVID-19) pandemic, determining antibody reactivity to SARS-CoV-2 epitopes becomes essential in discerning changes in the immune response to infection over time. This study enabled us to identify specific areas of antigenicity within the SARS-CoV-2 proteome, allowing us to detect correlations of epitopes with clinical metadata and immunological signals to gain holistic insight into SARS-CoV-2 infection. This work also emphasized the risk of mutation accumulation in viral variants and the potential for evasion of the adaptive immune responses in the event of reinfection. We additionally highlighted the correlation of antigenicity between structural proteins of SARS-CoV-2 and endemic HCoVs, raising the possibility of cross-protection between homologous lineages. Finally, we identified a subset of patients with minimal antibody reactivity to SARS-CoV-2 infection, prompting discussion of the potential consequences of this alternative immune response.
Keywords: SARS-CoV-2; clinical severity; endemic coronaviruses; epitopes; humoral immunity; viral variants.
Figures



References
-
- Ren L, Fan G, Wu W, Guo L, Wang Y, Li X, Wang C, Gu X, Li C, Wang Y, Wang G, Zhou F, Liu Z, Ge Q, Zhang Y, Li H, Zhang L, Xu J, Wang C, Wang J, Cao B. 2021. Antibody responses and clinical outcomes in adults hospitalized with severe coronavirus disease 2019 (COVID-19): a post hoc analysis of LOTUS China trial. Clin Infect Dis 72:e545–e551. doi:10.1093/cid/ciaa1247. - DOI - PMC - PubMed
-
- Lynch KL, Whitman JD, Lacanienta NP, Beckerdite EW, Kastner SA, Shy BR, Goldgof GM, Levine AG, Bapat SP, Stramer SL, Esensten JH, Hightower AW, Bern C, Wu AHB. 2021. Magnitude and kinetics of anti-severe acute respiratory syndrome coronavirus 2 antibody responses and their relationship to disease severity. Clin Infect Dis 72:301–308. doi:10.1093/cid/ciaa979. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

