Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative
- PMID: 34183353
- PMCID: PMC9414283
- DOI: 10.1158/2159-8290.CD-20-1647
Genomic and Transcriptomic Analyses of Breast Cancer Primaries and Matched Metastases in AURORA, the Breast International Group (BIG) Molecular Screening Initiative
Abstract
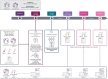
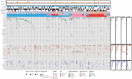
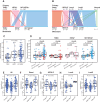
AURORA aims to study the processes of relapse in metastatic breast cancer (MBC) by performing multi-omics profiling on paired primary tumors and early-course metastases. Among 381 patients (primary tumor and metastasis pairs: 252 targeted gene sequencing, 152 RNA sequencing, 67 single nucleotide polymorphism arrays), we found a driver role for GATA1 and MEN1 somatic mutations. Metastases were enriched in ESR1, PTEN, CDH1, PIK3CA, and RB1 mutations; MDM4 and MYC amplifications; and ARID1A deletions. An increase in clonality was observed in driver genes such as ERBB2 and RB1. Intrinsic subtype switching occurred in 36% of cases. Luminal A/B to HER2-enriched switching was associated with TP53 and/or PIK3CA mutations. Metastases had lower immune score and increased immune-permissive cells. High tumor mutational burden correlated to shorter time to relapse in HR+/HER2- cancers. ESCAT tier I/II alterations were detected in 51% of patients and matched therapy was used in 7%. Integration of multi-omics analyses in clinical practice could affect treatment strategies in MBC. SIGNIFICANCE: The AURORA program, through the genomic and transcriptomic analyses of matched primary and metastatic samples from 381 patients with breast cancer, coupled with prospectively collected clinical data, identified genomic alterations enriched in metastases and prognostic biomarkers. ESCAT tier I/II alterations were detected in more than half of the patients.This article is highlighted in the In This Issue feature, p. 2659.
©2021 The Authors; Published by the American Association for Cancer Research.
Figures



Comment in
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

