Genetic modifiers regulating DNA replication and double-strand break repair are associated with differences in mammary tumors in mouse models of Li-Fraumeni syndrome
- PMID: 34183771
- PMCID: PMC8349885
- DOI: 10.1038/s41388-021-01892-5
Genetic modifiers regulating DNA replication and double-strand break repair are associated with differences in mammary tumors in mouse models of Li-Fraumeni syndrome
Abstract
Breast cancer is the most common tumor among women with inherited variants in the TP53 tumor suppressor, but onset varies widely suggesting interactions with genetic or environmental factors. Rodent models haploinsufficent for Trp53 also develop a wide variety of malignancies associated with Li-Fraumeni syndrome, but BALB/c mice are uniquely susceptible to mammary tumors and is genetically linked to the Suprmam1 locus on chromosome 7. To define mechanisms that interact with deficiencies in p53 to alter susceptibility to mammary tumors, we fine mapped the Suprmam1 locus in females from an N2 backcross of BALB/cMed and C57BL/6J mice. A major modifier was localized within a 10 cM interval on chromosome 7. The effect of the locus on DNA damage responses was examined in the parental strains and mice that are congenic for C57BL/6J alleles on the BALB/cMed background (SM1-Trp53+/-). The mammary epithelium of C57BL/6J-Trp53+/- females exhibited little radiation-induced apoptosis compared to BALB/cMed-Trp53+/- and SM1-Trp53+/- females indicating that the Suprmam1B6/B6 alleles could not rescue repair of radiation-induced DNA double-strand breaks mostly relying on non-homologous end joining. In contrast, the Suprmam1B6/B6 alleles in SM1-Trp53+/- mice were sufficient to confer the C57BL/6J-Trp53+/- phenotypes in homology-directed repair and replication fork progression. The Suprmam1B6/B6 alleles in SM1-Trp53+/- mice appear to act in trans to regulate a panel of DNA repair and replication genes which lie outside the locus.
© 2021. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
The authors declare no potential conflicts of interest.
Figures

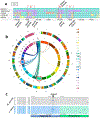
 ) and subsequent strains are assigned the same color where their haplotypes match the first strain i.e. 129S1SvimJ alleles that differed from BALB/c (
) and subsequent strains are assigned the same color where their haplotypes match the first strain i.e. 129S1SvimJ alleles that differed from BALB/c ( ), FVB/NJ alleles different from BALB/cJ and FVB/NJ (
), FVB/NJ alleles different from BALB/cJ and FVB/NJ ( ), C57BL/6 that different from 3 previous tracks (
), C57BL/6 that different from 3 previous tracks ( ) and DBA2/J alleles that is different from all 4 track above (
) and DBA2/J alleles that is different from all 4 track above ( ). Region A (125-127 Mb) defines a block that is within the shoulder region of linkage bounded by polymorphisms in Plekha7 and Abca14 in Fig. 1a. Regions B (132-133.2 Mb) and C (134.5-136 Mb) lie within the peak linkage and flank Rabep2. b) Map of human chromosomes syntenic with mouse chromosome 7. The Synteny Browser was used to define relationships between mouse and human chromosomes [54]. The outer ring represents mouse chromsomes, the inner ring represents human chromsomes and the connections show syntenic regions to mouse chromosome 7. Regions of synteny to Suprmam1 interval shown with red circle in the human chromosome 10 and 16. The color key represents human chromsomes with the non-syntenic chromosomes shaded. c) Expaned view of Suprmam1 syntenic regions. The peak linkage surrounding the polymorphism in Rabep2 is syntenic with human chromosome 16. The more telomeric region of the Suprmam1 interval is syntenic with human chromosome 10. Grey bars indicate locations of genes.
). Region A (125-127 Mb) defines a block that is within the shoulder region of linkage bounded by polymorphisms in Plekha7 and Abca14 in Fig. 1a. Regions B (132-133.2 Mb) and C (134.5-136 Mb) lie within the peak linkage and flank Rabep2. b) Map of human chromosomes syntenic with mouse chromosome 7. The Synteny Browser was used to define relationships between mouse and human chromosomes [54]. The outer ring represents mouse chromsomes, the inner ring represents human chromsomes and the connections show syntenic regions to mouse chromosome 7. Regions of synteny to Suprmam1 interval shown with red circle in the human chromosome 10 and 16. The color key represents human chromsomes with the non-syntenic chromosomes shaded. c) Expaned view of Suprmam1 syntenic regions. The peak linkage surrounding the polymorphism in Rabep2 is syntenic with human chromosome 16. The more telomeric region of the Suprmam1 interval is syntenic with human chromosome 10. Grey bars indicate locations of genes.



References
-
- Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J et al. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat 2016; 37: 865–876. - PubMed
-
- Amadou A, Achatz MIW, Hainaut P. Revisiting tumor patterns and penetrance in germline TP53 mutation carriers: temporal phases of Li-Fraumeni syndrome. Curr Opin Oncol 2018; 30: 23–29. - PubMed
-
- Fortuno C, James PA, Spurdle AB. Current review of TP53 pathogenic germline variants in breast cancer patients outside Li-Fraumeni syndrome. Human mutation 2018; 39: 1764–1773. - PubMed
-
- Wendt C, Margolin S. Identifying breast cancer susceptibility genes - a review of the genetic background in familial breast cancer. Acta oncologica (Stockholm, Sweden) 2019; 58: 135–146. - PubMed
-
- Milne RL, Antoniou AC. Modifiers of breast and ovarian cancer risks for BRCA1 and BRCA2 mutation carriers. Endocr Relat Cancer 2016; 23: T69–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

